Band Structure Energy Shifted by 0.8 eV
Posted: Wed Dec 21, 2011 4:29 pm
Dear All,
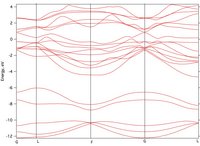
I have performed Band Structure Calculations on Sb2Te3 but although the main features compare well with published data and a castep calculation, the energy of all bands appears to be shifted up by 0.8 eV and I do not understand the reason. I have attached the bandstructure graphs from Castep and from Abinit to this post.
My procedure is as follows:
-Fully relax the atomic positions and cell geometry.
-Enter the relaxed cell structure into a new file for the band structure calculation.
-Do a single point energy calculation to generate the wavefunctions.
-Get the electron density from the previous calculation and do a non self consistent calculation
During the Band Structure calculation I do receive a few warnings regarding the hdr check. Specifcally:
-P-0000 hdr_check: WARNING -
-P-0000 input nkpt= 190 not equal disk file nkpt= 5
-P-0000 symafm:
and
-P-0000 hdr_check: WARNING -
-P-0000 Restart of self-consistent calculation need translated wavefunctions.
-P-0000 Indeed, critical differences between current calculation and
-P-0000 restart file have been detected in:
-P-0000 * the number, position, or weight of k-points
-P-0000 * the format of wavefunctions (istwfk)
I would be grateful if somebody could explain where I am making a mistake and how I can resolve the hdr_check warnings. I guess that my problem is related to these warnings.
Thanks,
Rob
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
I am running:
.Version 6.8.1 of ABINIT
.(MPI version, prepared for a x86_64_linux_gnu4.4 computer)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
My i/p file for relaxing the structure is:
ndtset 2 # atom position and geom
ionmov 2 # move atoms
optcell1 0
optcell2 2
getxred -1
getwfk -1
ntime 500 # Maximum number of optimization steps
nstep 200
dilatmx 1.05 # Maximum scaling allowed for lattice parameters
ecutsm 0.5 # Energy CUToff SMearing
# Structure
spgroup 166 #Space Group (1=p1)
spgaxor 2
natom 5
natrd 3
ntypat 2 #number of types of Atoms
znucl 51 52 #Z-number of atom types Sb51, Te52
typat 1 2 2
acell 10.60650792 10.60650792 10.60650792 angstroms
angdeg 23.48953218 23.48953218 23.48953218
xred
0.602824 0.602824 0.602824
0.785568 0.785568 0.785568
0 0 0
kptrlen 200
brvltt -1
diemac 12.0
ecut 36 #from the convergence study
ixc 11 #GGA
nband 20
nbdbuf 2
tolmxf 5.0d-9
toldff 5.0d-10
occopt 3 # Smearing of Bands
tsmear 0.001 Ha # Temperature of Smearing, 0.001 Ha = 27.2113845 meV = 315.773 Kelvin
prtcif 1
prtvol 2
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
My Input file for calculating the band structure is:
ndtset 2
ionmov 0 #Single point energy calc
optcell 0
getxred2 -1
getwfk2 -1
ecutsm 0.5
iscf 7
npulayit 10
nstep1 100
nstep2 500
prtden 1 getden1 0 getwfk1 0 ! Usual file handling data
prtcif 1
prtdos 1
prtvol 2
#Data common to all datasets
###### STRUCTURE #########
spgroup 166 #Space Group (1=p1)
spgaxor 2 #Rhombohedral Lattice 2
natom 5
natrd 3
ntypat 2 #number of types of Atoms
znucl 51 52 #Z-number of atom types Sb51, Te52
typat 1 2 2
acell 10.60766 10.60766 10.60766 angstroms
angdeg 23.486 23.486 23.486
xred
0.602828 0.602828 0.602828
0.785558 0.785558 0.785558
0 0 0
###### K points #########
kptopt1 1
kptopt2 -4
ngkpt 10 10 10
nshiftk 1
iscf2 -2
ndivk2 10 27 27 27
kptbounds2
0.0 0.0 0.0 # G point
0.5 0.5 0.5 # Z point
0.5 0.5 0.0 # F point
0.0 0.0 0.0 # G point
0.0 0.5 0.0 # L point
brvltt -1
diemac 12.0
ecut 36
ixc 11
#tnons 72*0.0
#### Tolerances #####
tolwfr2 1.e-24
tolmxf 5.0d-9
toldff 5.0d-10
##### BANDS ######
nband 20
nbdbuf 2
occopt 3 # Smearing of Bands
tsmear 0.001 Ha # Temperature of Smearing, 0.001 Ha = 27.2113845 meV = 315.773 Kelvin
getden2 -1
enunit2 1
I have performed Band Structure Calculations on Sb2Te3 but although the main features compare well with published data and a castep calculation, the energy of all bands appears to be shifted up by 0.8 eV and I do not understand the reason. I have attached the bandstructure graphs from Castep and from Abinit to this post.
My procedure is as follows:
-Fully relax the atomic positions and cell geometry.
-Enter the relaxed cell structure into a new file for the band structure calculation.
-Do a single point energy calculation to generate the wavefunctions.
-Get the electron density from the previous calculation and do a non self consistent calculation
During the Band Structure calculation I do receive a few warnings regarding the hdr check. Specifcally:
-P-0000 hdr_check: WARNING -
-P-0000 input nkpt= 190 not equal disk file nkpt= 5
-P-0000 symafm:
and
-P-0000 hdr_check: WARNING -
-P-0000 Restart of self-consistent calculation need translated wavefunctions.
-P-0000 Indeed, critical differences between current calculation and
-P-0000 restart file have been detected in:
-P-0000 * the number, position, or weight of k-points
-P-0000 * the format of wavefunctions (istwfk)
I would be grateful if somebody could explain where I am making a mistake and how I can resolve the hdr_check warnings. I guess that my problem is related to these warnings.
Thanks,
Rob
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
I am running:
.Version 6.8.1 of ABINIT
.(MPI version, prepared for a x86_64_linux_gnu4.4 computer)
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
My i/p file for relaxing the structure is:
ndtset 2 # atom position and geom
ionmov 2 # move atoms
optcell1 0
optcell2 2
getxred -1
getwfk -1
ntime 500 # Maximum number of optimization steps
nstep 200
dilatmx 1.05 # Maximum scaling allowed for lattice parameters
ecutsm 0.5 # Energy CUToff SMearing
# Structure
spgroup 166 #Space Group (1=p1)
spgaxor 2
natom 5
natrd 3
ntypat 2 #number of types of Atoms
znucl 51 52 #Z-number of atom types Sb51, Te52
typat 1 2 2
acell 10.60650792 10.60650792 10.60650792 angstroms
angdeg 23.48953218 23.48953218 23.48953218
xred
0.602824 0.602824 0.602824
0.785568 0.785568 0.785568
0 0 0
kptrlen 200
brvltt -1
diemac 12.0
ecut 36 #from the convergence study
ixc 11 #GGA
nband 20
nbdbuf 2
tolmxf 5.0d-9
toldff 5.0d-10
occopt 3 # Smearing of Bands
tsmear 0.001 Ha # Temperature of Smearing, 0.001 Ha = 27.2113845 meV = 315.773 Kelvin
prtcif 1
prtvol 2
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
My Input file for calculating the band structure is:
ndtset 2
ionmov 0 #Single point energy calc
optcell 0
getxred2 -1
getwfk2 -1
ecutsm 0.5
iscf 7
npulayit 10
nstep1 100
nstep2 500
prtden 1 getden1 0 getwfk1 0 ! Usual file handling data
prtcif 1
prtdos 1
prtvol 2
#Data common to all datasets
###### STRUCTURE #########
spgroup 166 #Space Group (1=p1)
spgaxor 2 #Rhombohedral Lattice 2
natom 5
natrd 3
ntypat 2 #number of types of Atoms
znucl 51 52 #Z-number of atom types Sb51, Te52
typat 1 2 2
acell 10.60766 10.60766 10.60766 angstroms
angdeg 23.486 23.486 23.486
xred
0.602828 0.602828 0.602828
0.785558 0.785558 0.785558
0 0 0
###### K points #########
kptopt1 1
kptopt2 -4
ngkpt 10 10 10
nshiftk 1
iscf2 -2
ndivk2 10 27 27 27
kptbounds2
0.0 0.0 0.0 # G point
0.5 0.5 0.5 # Z point
0.5 0.5 0.0 # F point
0.0 0.0 0.0 # G point
0.0 0.5 0.0 # L point
brvltt -1
diemac 12.0
ecut 36
ixc 11
#tnons 72*0.0
#### Tolerances #####
tolwfr2 1.e-24
tolmxf 5.0d-9
toldff 5.0d-10
##### BANDS ######
nband 20
nbdbuf 2
occopt 3 # Smearing of Bands
tsmear 0.001 Ha # Temperature of Smearing, 0.001 Ha = 27.2113845 meV = 315.773 Kelvin
getden2 -1
enunit2 1