I'm a beginner for first principle calculation and abinit.
I'm trying to calculate the electronic structure of hexagonal YMnO3.
I prepared PAW data sets for Y, Mn, and O using atompaw package,
and the grounded state calculation results were good agree with the references
(cell size, atomic position, bandgap, magnetization, band structure,...).
However, in the DOSs calculation, I met some problems.
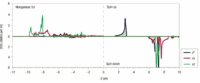
I hoped to calculate m-resolved density of states of Mn 3d for spin-up and down.
So I changed parameters : nsppol to 2 and wrote prtdos 3, prtdosm 1.
My result was:
and the reference:
ref:Nature Materials 3, 164 - 170 (2004)
http://www.nature.com/nmat/journal/v3/n ... t1080.html
For the up-spin state, it looks similar to the reference:
dxz,yz, dxy,x^2-y^2 doublets locates below the fermi leve, and dz^2 singlet above ~1eV from the fermi leve.
But for the down state, the reference and my result is quite different:
in my result, the DOSs of up and down were same except for the range near 4ev.
And the value of DOS were too large (above 400) compared to reference (2~3)
How to solve this problem? Is this problem originated from my PAW data sets?
Please help me
My input file and paw input file were
iscf 17
tolvrs 1.0d-10
kptopt 1
ngkpt 4 4 4
nshiftk 1
shiftk 0.0 0.0 0.0
prtdos 3
prtdosm 1
enunit 1
#Spin
nsppol 2
nspden 2
nspinor 1
spinat 0 0 0
0 0 0
0 0 3
0 0 -3
0 0 0
0 0 0
0 0 0
0 0 0
0 0 0
0 0 0
nstep 3000
ecut 15
pawecutdg 30
natom 10
ntypat 3
znucl 39 25 8
typat 1 1 2 2 3 3 3 3 3 3
angdeg
90 90 120
acell
6.8602016086E+00 6.8602016086E+00 2.1611255195E+01 Bohr
xred
-1.2766932425E-16 -6.8681497753E-16 -1.4981797821E-17
-1.2766932425E-16 -6.8681497753E-16 5.0000000000E-01
3.3333333333E-01 6.6666666667E-01 2.5000000000E-01
6.6666666667E-01 3.3333333333E-01 7.5000000000E-01
-1.2766932425E-16 -6.8681497753E-16 2.5000000000E-01
-1.2766932425E-16 -6.8681497753E-16 7.5000000000E-01
3.3333333333E-01 6.6666666667E-01 8.3935675655E-02
3.3333333333E-01 6.6666666667E-01 4.1606432434E-01
6.6666666667E-01 3.3333333333E-01 5.8393567566E-01
6.6666666667E-01 3.3333333333E-01 9.1606432434E-01
# LDA+U
usepawu 1
lpawu -1 2 -1
upawu 0.0 8.0 0.0 eV
jpawu 0.0 0.88 0.0 eV
usedmatpu 5
dmatpawu
1.00000 0.00000 0.00000 0.00000 0.00000
0.00000 1.00000 0.00000 0.00000 0.00000
0.00000 0.00000 1.00000 0.00000 0.00000
0.00000 0.00000 0.00000 1.00000 0.00000
0.00000 0.00000 0.00000 0.00000 1.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
0.00000 0.00000 0.00000 0.00000 0.00000
1.00000 0.00000 0.00000 0.00000 0.00000
0.00000 1.00000 0.00000 0.00000 0.00000
0.00000 0.00000 1.00000 0.00000 0.00000
0.00000 0.00000 0.00000 1.00000 0.00000
0.00000 0.00000 0.00000 0.00000 1.00000
<MN>
Mn 25
GGA-PBE loggrid 2001
4 4 3 0 0 0
3 2 6
4 0 1
4 1 0
0 0 0
c
c
v
v
c
v
v
v
2
1.9 1.24 1.9 1.9
n
n
y
2
n
custom polynom2 8 10
3 0 ultrasoft !troulliermartins
1.9
1.9
1.9
1.9
1.9
1.9
2
usexcnhat
0
<Y>
Y 39
GGA-PBE loggrid 2001
5 5 4 0 0 0
4 2 1
5 0 2
5 1 0
0 0 0
c
c
c
v
v
c
c
v
v
c
v
2
2.0
n
n
y
3
n
custom polynom2 8 10
3 0 ultrasoft
2.0
2.0
2.0
2.0
2.0
2.0
2
usexcnhat
0
<O>
O 8
GGA-PBE loggrid 1201
2 2 0 0 0 0
2 1 4
0 0 0
c
v
v
1
1.45
y
-0.676
n
y
-1.742
n
custom polynom2 8 10
2 0 ultrasoft
1.45
1.45
1.45
1.45
1
2 1 4
0 0 0
2
usexcnhat
0