Input file for cubic fcc crystal structure
Moderator: bguster
-
Mukul.pu02
- Posts: 17
- Joined: Sat Apr 23, 2011 9:34 am
Input file for cubic fcc crystal structure
Hello everyone
I want to make an input file to find out the total energy of a cubic fcc crystal structure x2y2z5 consisting of total 27 atoms with 3 vacancies.Please guide me:
! . what is the required command to generate supercell?
2. What is the method to generate the atomic positions for the crystal structure?
I will be highly grateful for your kind consideration
I want to make an input file to find out the total energy of a cubic fcc crystal structure x2y2z5 consisting of total 27 atoms with 3 vacancies.Please guide me:
! . what is the required command to generate supercell?
2. What is the method to generate the atomic positions for the crystal structure?
I will be highly grateful for your kind consideration
Re: Input file for cubic fcc crystal structure
Hi,
There is no supercell specific command. If you have a simple cubic crystal, you would have :
rprim
1 0 0
0 1 0
0 0 1
natom 1
typat 1
xred
0 0 0
Instad, if you use a 2*2*2 supercell with one vacancy, you'll have :
rprim
2 0 0
0 2 0
0 0 2
natom 7 #8 atoms and 1 vacancy
typat 7*1
xred
0 0 0
1 0 0
#0 1 0 #The vacancy
1 1 0
0 0 1
1 0 1
0 1 0
1 1 1
Also, don't forget that a supercell with a volume N time that of the primitive cell has a Brillouin's zone N time smaller. Therefore, you can reduce the number of k-points of your calculation by a factor N and keep the same precision.
Finally, if your supercell is a true supercell without any defect, use :
chkprim 0
The atomic positions in the supercell must all be written by the user in the input file. Abinit does not automatically compute them from the primitive cell.
1. what is the required command to generate supercell?
There is no supercell specific command. If you have a simple cubic crystal, you would have :
rprim
1 0 0
0 1 0
0 0 1
natom 1
typat 1
xred
0 0 0
Instad, if you use a 2*2*2 supercell with one vacancy, you'll have :
rprim
2 0 0
0 2 0
0 0 2
natom 7 #8 atoms and 1 vacancy
typat 7*1
xred
0 0 0
1 0 0
#0 1 0 #The vacancy
1 1 0
0 0 1
1 0 1
0 1 0
1 1 1
Also, don't forget that a supercell with a volume N time that of the primitive cell has a Brillouin's zone N time smaller. Therefore, you can reduce the number of k-points of your calculation by a factor N and keep the same precision.
Finally, if your supercell is a true supercell without any defect, use :
chkprim 0
2. What is the method to generate the atomic positions for the crystal structure?
The atomic positions in the supercell must all be written by the user in the input file. Abinit does not automatically compute them from the primitive cell.
Re: Input file for cubic fcc crystal structure
Hi,
It seems the geometry builder can be used to do just what you asked, sorry for the inaccuracy of my first reply. To create the supercell, you will need to define your rprim and acell according to the supercell you want. In my case, I want the 2*2*2 supercell of a simple cubic crystal of periodicity 2.67 bohr :
Don't forget to set chkprim to 0. Then, you define the atomic coordinates of the unit cell you usually work with. In my case, I had to work with a doubled cell (in z direction), since the geometry builder refuses to work with only one atom. Hence :
and I specify that the parser has only 2 atomic positions to read from the input by specifying:
Now, to have the 2*2*2 supercell, I need to copy the 2 atoms in xred one time 2.67 bohr away in the x direction. Then, I need to copy the 4 resulting atom one time 2.67 bohr away in the y direction. This is done by defining one geometry builder object:
applying to two atoms:
namely, atoms 1 and 2 in the list of given atoms (xred):
We want this object to produce a set of coordinates containing the set of atoms given in xred twice in the x direction, twice in the y direction and one time in the z direction:
The replicas will be 2.67 bohr distant in the x and y directions:
FInally, you need to adjust the number of atoms and the list of types of atoms to describe the total set of atoms generated by the geometry builder. In my case:
However, if you want to add vacancies, you simply need to define the variables vacunum and vaclst. I my case, I want one vacancy:
and it is the first atom in the list of 8 atoms generated by the geometry builder:
The vacancies need to be removed from the number of atoms and the list giving the type of each one:
More details on each keyword are given in the Abinit website :
http://www.abinit.org/documentation/helpfiles/for-v6.8/input_variables/vargeo.html
Here is an input file that works for me, with abinit 6.8.2 (it's the input resulting from my cubic example):
Good luck!
It seems the geometry builder can be used to do just what you asked, sorry for the inaccuracy of my first reply. To create the supercell, you will need to define your rprim and acell according to the supercell you want. In my case, I want the 2*2*2 supercell of a simple cubic crystal of periodicity 2.67 bohr :
Code: Select all
acell 3*5.34
rprim 1 0 0 0 1 0 0 0 1
chkprim 0Don't forget to set chkprim to 0. Then, you define the atomic coordinates of the unit cell you usually work with. In my case, I had to work with a doubled cell (in z direction), since the geometry builder refuses to work with only one atom. Hence :
Code: Select all
xred
0 0 0
0 0 1/2and I specify that the parser has only 2 atomic positions to read from the input by specifying:
Code: Select all
natrd 2Now, to have the 2*2*2 supercell, I need to copy the 2 atoms in xred one time 2.67 bohr away in the x direction. Then, I need to copy the 4 resulting atom one time 2.67 bohr away in the y direction. This is done by defining one geometry builder object:
Code: Select all
nobj 1applying to two atoms:
Code: Select all
objan 2namely, atoms 1 and 2 in the list of given atoms (xred):
Code: Select all
objaat 1 2We want this object to produce a set of coordinates containing the set of atoms given in xred twice in the x direction, twice in the y direction and one time in the z direction:
Code: Select all
objarf 2 2 1The replicas will be 2.67 bohr distant in the x and y directions:
Code: Select all
objatr
0.0 0.0 0.0
2.67 0.0 0.0
0.0 2.67 0.0
0.0 0.0 0.0FInally, you need to adjust the number of atoms and the list of types of atoms to describe the total set of atoms generated by the geometry builder. In my case:
Code: Select all
natom 8
typat 8*1However, if you want to add vacancies, you simply need to define the variables vacunum and vaclst. I my case, I want one vacancy:
Code: Select all
vacnum 1and it is the first atom in the list of 8 atoms generated by the geometry builder:
Code: Select all
vaclst 1The vacancies need to be removed from the number of atoms and the list giving the type of each one:
Code: Select all
natom 7
typat 7*1More details on each keyword are given in the Abinit website :
http://www.abinit.org/documentation/helpfiles/for-v6.8/input_variables/vargeo.html
Here is an input file that works for me, with abinit 6.8.2 (it's the input resulting from my cubic example):
Code: Select all
# Test file for Geometry builder
#Definition of the unit cell
acell 3*5.34
rprim 1 0 0 0 1 0 0 0 1
chkprim 0
natrd 2
nobj 1
objaat 1 2
objan 2
objarf 2 2 1
objatr
0.0 0.0 0.0
2.67 0.0 0.0
0.0 2.67 0.0
0.0 0.0 0.0
vacnum 1
vaclst 1
#Definition of the atom types
ntypat 1
znucl 6
#Definition of the atoms
natom 7
typat 7*1
xred
0 0 0
0 0 1/2
#Definition of the planewave basis set
ecut 10.0
#Definition of the SCF procedure
nstep 100
diemac 2.0
# add to conserve old < 6.7.2 behavior for calculating forces at each SCF step
optforces 1
# k-points grid
kptopt 1
nshiftk 1
shiftk 0.0 0.0 0.0
ngkpt 2 2 2
toldfe 1.0d-10Good luck!
-
Mukul.pu02
- Posts: 17
- Joined: Sat Apr 23, 2011 9:34 am
Re: Input file for cubic fcc crystal structure
Hello jolafc,
First of all thank you very much for your response.It has really helped to understand these basic input variables which i found difficult initially.
I have tried to make my input file.
would you please guide me how can we introduce vacancy with the mentioned input variable because i got some errors.
Please guide.
First of all thank you very much for your response.It has really helped to understand these basic input variables which i found difficult initially.
I have tried to make my input file.
would you please guide me how can we introduce vacancy with the mentioned input variable because i got some errors.
Please guide.
Re: Input file for cubic fcc crystal structure
Hi Mukul.pu02,
It is difficult to address all of the different problems in one reply. If you could post your input file, the pseudopotentials you use and copy-paste the error message you get in your log file when you run your calculation, I would be pleased to help you.
It is difficult to address all of the different problems in one reply. If you could post your input file, the pseudopotentials you use and copy-paste the error message you get in your log file when you run your calculation, I would be pleased to help you.
-
Mukul.pu02
- Posts: 17
- Joined: Sat Apr 23, 2011 9:34 am
Re: Input file for cubic fcc crystal structure
Hello jolafc,
Thank you very much once again for your kind consideration.First of all i would like to tell you that i want to make a 2*2*2 supercell consisting of total 64 atoms having vacancies positions.The input file is attached for reference.
Secondly,the 3 pseudo potential files which i have been using in my case are: 32ge.pspnc , 51sb.pspnc , 52te.pspnc.Sorry,I am unable to upload them this time because there is problem that the extension is not allowed.
The log file indicate the following error:
"ingeobld : ERROR --
The number of atoms to be read (natrd) must be equal
to the total number of atoms (natom), plus
the number of vacancies (vacnum), minus
the number of atoms added by the repetition of objects.
This is not the case : natrd= 12, natom= 64, vacnum= 1;
object a : objan= 8, objarf(1:3)= 2 2 2 => adds 56 atoms.
object b : objbn= 4, objbrf(1:3)= 2 2 2 => adds 28 atoms.
Action : check the correspondence between natom+vacnum on one side,
and natrd, objan, objbn, objarf and objbrf on the other side. "
Please guide.Waiting for your response.
Regards
Thank you very much once again for your kind consideration.First of all i would like to tell you that i want to make a 2*2*2 supercell consisting of total 64 atoms having vacancies positions.The input file is attached for reference.
Secondly,the 3 pseudo potential files which i have been using in my case are: 32ge.pspnc , 51sb.pspnc , 52te.pspnc.Sorry,I am unable to upload them this time because there is problem that the extension is not allowed.
The log file indicate the following error:
"ingeobld : ERROR --
The number of atoms to be read (natrd) must be equal
to the total number of atoms (natom), plus
the number of vacancies (vacnum), minus
the number of atoms added by the repetition of objects.
This is not the case : natrd= 12, natom= 64, vacnum= 1;
object a : objan= 8, objarf(1:3)= 2 2 2 => adds 56 atoms.
object b : objbn= 4, objbrf(1:3)= 2 2 2 => adds 28 atoms.
Action : check the correspondence between natom+vacnum on one side,
and natrd, objan, objbn, objarf and objbrf on the other side. "
Please guide.Waiting for your response.
Regards
- Attachments
-
 s23.in
s23.in- Input File
- (1.34 KiB) Downloaded 475 times
Re: Input file for cubic fcc crystal structure
Here is a diff showing how to correct your input :
diff s23.bak s23.in
Since you translate your atoms by 5.68 bohr, your supercell HAS to be 2*5.68 bohr big. And, your prim*acell vector must define your supercell, not your primitive cell. Therefore, if you use acell 11.36 11.36 11.36 bohr, rprim has to be 1 0 0 0 1 0 0 0 1.
Since you apply the same translations operations on all your atoms, you just have to define a single object, not 2.
You misinterpreted the variable natom. It represents the final number of atoms in your supercell, after all replications of atoms and subtractions of vacancies have been done. Here, you repeat 8 times your primitive cell of 12 atoms and subtract one vacancy afterwards, meaning natom = 8*12-1=95.
Also, typat needs to be defined for the atoms appearing in xred only, not all atoms of the supercell. Hence, you only need to give 12 numbers.
Since I redefined your cell as the supercell by doubling rprim, I needed to halve the coordinates of the atoms in the "primitive" basis (defined as the product acell*rprim, which here is in fact your supercell).
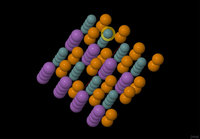
The resulting input passes in abinit 6.8.2. It gives the structure given in attachment, with the highlighted atom in fact being replaced by a vacancy.
Good luck!
diff s23.bak s23.in
Code: Select all
5c5
< rprim 0.5 0 0 0 0.5 0 0 0 0.5
---
> rprim 1 0 0 0 1 0 0 0 1
Since you translate your atoms by 5.68 bohr, your supercell HAS to be 2*5.68 bohr big. And, your prim*acell vector must define your supercell, not your primitive cell. Therefore, if you use acell 11.36 11.36 11.36 bohr, rprim has to be 1 0 0 0 1 0 0 0 1.
Code: Select all
9,13c9,11
< nobj 2
< objaat 1 2 3 4 5 6 7 8
< objbat 9 10 11 12
< objan 8
< objbn 4
---
> nobj 1
> objaat 1 2 3 4 5 6 7 8 9 10 11 12
> objan 12
15d12
< objbrf 2 2 2
21,25d17
< objbtr
< 0.0 0.0 0.0
< 5.68 0.0 0.0
< 0.0 5.68 0.0
< 0.0 0.0 5.68
Since you apply the same translations operations on all your atoms, you just have to define a single object, not 2.
Code: Select all
36,37c28,29
< natom 64
< typat 16*1 16*2 32*3
---
> natom 95
> typat 4*1 4*2 4*3
You misinterpreted the variable natom. It represents the final number of atoms in your supercell, after all replications of atoms and subtractions of vacancies have been done. Here, you repeat 8 times your primitive cell of 12 atoms and subtract one vacancy afterwards, meaning natom = 8*12-1=95.
Also, typat needs to be defined for the atoms appearing in xred only, not all atoms of the supercell. Hence, you only need to give 12 numbers.
Code: Select all
39,46c31,38
< 0.25 0.25 0.25 #ge
< 0.25 0.25 0.75 #ge
< 0.25 0.75 0.25#-v-
< 0.25 0.75 0.75#ge
< 0.75 0.25 0.25#sb
< 0.75 0.75 0.25#sb
< 0.75 0.25 0.75#sb
< 0.75 0.75 0.75 # sb
---
> 0.125 0.125 0.125 #ge
> 0.125 0.125 0.375 #ge
> 0.125 0.375 0.125#-v-
> 0.125 0.375 0.375#ge
> 0.375 0.125 0.125#sb
> 0.375 0.375 0.125#sb
> 0.375 0.125 0.375#sb
> 0.375 0.375 0.375 # sb
48,50c40,42
< 0.5 0 0.5#te
< 0.5 0.5 0#te
< 0 0.5 0.5 #te
---
> 0.25 0 0.5#te
> 0.25 0.25 0#te
> 0 0.25 0.25 #te
Since I redefined your cell as the supercell by doubling rprim, I needed to halve the coordinates of the atoms in the "primitive" basis (defined as the product acell*rprim, which here is in fact your supercell).
The resulting input passes in abinit 6.8.2. It gives the structure given in attachment, with the highlighted atom in fact being replaced by a vacancy.
Good luck!
-
Mukul.pu02
- Posts: 17
- Joined: Sat Apr 23, 2011 9:34 am
Re: Input file for cubic fcc crystal structure
Hello Sir
Thanks a lot for this response.I am correcting my input file as per your guidance.
Sir you have attached a image file of crystal structure so formed.
1) Please tell me what does the gap between two purple coloured atoms show since the vacancy position is shown through the highlighted atom.
2) Kindly tell me the name of software used to view the required crystal structure.
Kind Regards
M.Mukul
Thanks a lot for this response.I am correcting my input file as per your guidance.
Sir you have attached a image file of crystal structure so formed.
1) Please tell me what does the gap between two purple coloured atoms show since the vacancy position is shown through the highlighted atom.
2) Kindly tell me the name of software used to view the required crystal structure.
Kind Regards
M.Mukul
Re: Input file for cubic fcc crystal structure
Hi!
First, don't blindly copy my corrections to your input file. I just saw that I wrote
for one of the Te atom, while the real coordinates are
Also, I don't understand what you mean by "what does the gap between two purple coloured atoms show"... Maybe you could illustrate your question?
About illustrating and visualising a crystal structure, I use 2 things to produce a picture such as the one attached in my previous post. First, use the AbinitStructureViewer2.py file (attached at the end of this post with the .py extension replaced by .in to allow the file to be attached; replace it by .py to be able to use it) on one of you output :
This assumes that you have access to a shell (e.g. you run abinit on a mac or a linux computer) and that you have python installed somewhere with numpy available. It also assumes that you put the AbinitStructureViewer2.py file in your home directory (just change the path ~/AbinitStructureViewer2.py to something else if you put it elsewhere). Also, you need to be in the directory where the s23.out file is.
Once you have run the code, you should see :
You just mention the number of time you want the cell specified in your input to be replicated. To produce my picture, I just used 1 1 1. Then, you should see :
and have a new file in your folder named s23_outL.xyz.
You can use jmol to open such a file and visualize your structure in 3D, take pictures of it, etc. Jmol is available there :
http://jmol.sourceforge.net/download/
althought I just installed my version (on a mac) using macports (http://www.macports.org/) and it is also easy to use your favorite package manager in linux to install it.
If you could highlight the atoms sourrounding the gap you spoke about and post the picture, it would allow me to visualize your question.
Jonathan
First, don't blindly copy my corrections to your input file. I just saw that I wrote
Code: Select all
0.25 0 0.5#tefor one of the Te atom, while the real coordinates are
Code: Select all
0.25 0 0.25#teAlso, I don't understand what you mean by "what does the gap between two purple coloured atoms show"... Maybe you could illustrate your question?
About illustrating and visualising a crystal structure, I use 2 things to produce a picture such as the one attached in my previous post. First, use the AbinitStructureViewer2.py file (attached at the end of this post with the .py extension replaced by .in to allow the file to be attached; replace it by .py to be able to use it) on one of you output :
Code: Select all
python ~/AbinitStructureViewer2.py s23.outThis assumes that you have access to a shell (e.g. you run abinit on a mac or a linux computer) and that you have python installed somewhere with numpy available. It also assumes that you put the AbinitStructureViewer2.py file in your home directory (just change the path ~/AbinitStructureViewer2.py to something else if you put it elsewhere). Also, you need to be in the directory where the s23.out file is.
Once you have run the code, you should see :
Code: Select all
$ /opt/local/bin/python2.5 /Users/jonathan/programmes/AbinitStructureViewer2.py s23.outL
==========================
AbinitStructureViewer.py
version beta
==========================
- WARNING : rprim not fount in given file, assuming : rprim 1 0 0 0 1 0 0 0 1
1 configuration(s) found
- WARNING : calculation not completed in the given file -
Enter the number of times you wish to replicate the primitive cell
(If you only want the primitive cell, enter : 1 1 1)
You just mention the number of time you want the cell specified in your input to be replicated. To produce my picture, I just used 1 1 1. Then, you should see :
Code: Select all
"s23_outL.xyz" file created successfullyand have a new file in your folder named s23_outL.xyz.
You can use jmol to open such a file and visualize your structure in 3D, take pictures of it, etc. Jmol is available there :
http://jmol.sourceforge.net/download/
althought I just installed my version (on a mac) using macports (http://www.macports.org/) and it is also easy to use your favorite package manager in linux to install it.
If you could highlight the atoms sourrounding the gap you spoke about and post the picture, it would allow me to visualize your question.
Jonathan
- Attachments
-
 AbinitStructureViewer2.in
AbinitStructureViewer2.in- (28.6 KiB) Downloaded 465 times
-
Mukul.pu02
- Posts: 17
- Joined: Sat Apr 23, 2011 9:34 am
Re: Input file for cubic fcc crystal structure
Hello Sir,
i wanted to thank you for taking the time to reply to my problem in such a prompt manner.
Thank you so much for the help and advice you have shared with me in the past.
I got my last question's answer too through your last post.
BEST REGARDS
M.Mukul
i wanted to thank you for taking the time to reply to my problem in such a prompt manner.
Thank you so much for the help and advice you have shared with me in the past.
I got my last question's answer too through your last post.
BEST REGARDS
M.Mukul
-
Seba Darshan
- Posts: 63
- Joined: Mon Feb 17, 2014 5:19 am
Re: Input file for cubic fcc crystal structure
hai
I want to create a 2*2*2 supercell of graphene having a lattice constant of 4.6469 Bohr with single vacancy.
I would like to know how can I use the geometry builder to build the graphene sheet. I am not sure of how to use these variables. Can I give the geometry builder variables in the normal input file (filename.in file).
I managed to create a 1*1 unit cell of graphene without any error without using geometry builder input variables. . I am not getting a clear idea of how can I give the atomic positions in geometry builder.
How to change acell and rpriim to make it a 2*2 supercell. is it correct to use like this: acell 9.2988 9.2988 9.2988
rprim 0.866 0.5 0.0
-0.866 0.5 0.0
0.0 0.0 1.0. Whether this represents a 2*2*2 graphene supercell.
I also want to know how can I specify the position of the vacancy.
Regards
Seba
I want to create a 2*2*2 supercell of graphene having a lattice constant of 4.6469 Bohr with single vacancy.
I would like to know how can I use the geometry builder to build the graphene sheet. I am not sure of how to use these variables. Can I give the geometry builder variables in the normal input file (filename.in file).
I managed to create a 1*1 unit cell of graphene without any error without using geometry builder input variables. . I am not getting a clear idea of how can I give the atomic positions in geometry builder.
How to change acell and rpriim to make it a 2*2 supercell. is it correct to use like this: acell 9.2988 9.2988 9.2988
rprim 0.866 0.5 0.0
-0.866 0.5 0.0
0.0 0.0 1.0. Whether this represents a 2*2*2 graphene supercell.
I also want to know how can I specify the position of the vacancy.
Regards
Seba