polarization by lda+u and paw pseudopotention
Moderator: bguster
polarization by lda+u and paw pseudopotention
Hello,
i wana to use lda+u to calculate polarization of bifeo3.i have an WARNING like this.
could you please tell me why i have this warning?
input file:
acell 5.58132 5.58132 13.87698 angstrom
angdeg 90 90 120
spgroup 161
brvltt -1
chkprim 0
#Atom definition
natom 10
natrd 3
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
#SCF parameters
ixc 7
iscf 17
nstep 100
#planewave basis
ecut 35
pawecutdg 50
pawovlp -1.5
kptopt 4
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-12
#Berry phase calculation of the polarization
berryopt -1
rfdir 1 1 1
xred 0.000000000 0.000000000 0.000000000
0.000000000 0.000000000 0.221200004
0.442999989 0.012000000 0.954299986
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.9 0.0 ev
and WARNING:
chkpawovlp : WARNING -
PAW SPHERES ARE OVERLAPPING !
Distance between atoms 2 and 4 is : 3.66418
PAW radius of the sphere around atom 2 is: 2.60648
PAW radius of the sphere around atom 4 is: 2.31741
This leads to a (voluminal) overlap ratio of 10.71 %
COMPENSATION DENSITIES ARE OVERLAPPING !!!!
Distance between atoms 2 and 4 is : 3.66418
Compensation radius around atom 2 is: 2.60648
Compensation radius around atom 4 is: 2.01261
This leads to a (voluminal) overlap ratio of 8.82 %
THIS IS DANGEROUS !, as PAW formalism assume non-overlapping compensation densities.
=> Note that other spheres are overlapping !
Results might be approximate,
and even inaccurate (if overlap is too big) !
Assume experienced user. Execution will continue.
scfcv : before setvtr, energies%e_hartree= 0.000000000000000E+000
ewald : nr and ng are 4 and 10
setvtr : istep,n1xccc,moved_rhor= 1 1 0
scfcv : after setvtr, energies%e_hartree= 60.0094533400047
Thankyou.
~
"polnaf.in" 42L, 680C
i wana to use lda+u to calculate polarization of bifeo3.i have an WARNING like this.
could you please tell me why i have this warning?
input file:
acell 5.58132 5.58132 13.87698 angstrom
angdeg 90 90 120
spgroup 161
brvltt -1
chkprim 0
#Atom definition
natom 10
natrd 3
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
#SCF parameters
ixc 7
iscf 17
nstep 100
#planewave basis
ecut 35
pawecutdg 50
pawovlp -1.5
kptopt 4
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-12
#Berry phase calculation of the polarization
berryopt -1
rfdir 1 1 1
xred 0.000000000 0.000000000 0.000000000
0.000000000 0.000000000 0.221200004
0.442999989 0.012000000 0.954299986
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.9 0.0 ev
and WARNING:
chkpawovlp : WARNING -
PAW SPHERES ARE OVERLAPPING !
Distance between atoms 2 and 4 is : 3.66418
PAW radius of the sphere around atom 2 is: 2.60648
PAW radius of the sphere around atom 4 is: 2.31741
This leads to a (voluminal) overlap ratio of 10.71 %
COMPENSATION DENSITIES ARE OVERLAPPING !!!!
Distance between atoms 2 and 4 is : 3.66418
Compensation radius around atom 2 is: 2.60648
Compensation radius around atom 4 is: 2.01261
This leads to a (voluminal) overlap ratio of 8.82 %
THIS IS DANGEROUS !, as PAW formalism assume non-overlapping compensation densities.
=> Note that other spheres are overlapping !
Results might be approximate,
and even inaccurate (if overlap is too big) !
Assume experienced user. Execution will continue.
scfcv : before setvtr, energies%e_hartree= 0.000000000000000E+000
ewald : nr and ng are 4 and 10
setvtr : istep,n1xccc,moved_rhor= 1 1 0
scfcv : after setvtr, energies%e_hartree= 60.0094533400047
Thankyou.
~
"polnaf.in" 42L, 680C
Re: polarization by lda+u and paw pseudopotention
Your Bi and Fe PAW spheres are large enough that they are overlapping in real space. The PAW theory depends on the assumption that the PAW spheres do not overlap, therefore your calculation is highly suspect and probably will be wrong. You need to use PAW radii that are small enough that the spheres do not overlap. This may mean that you will have to make your own PAW data sets for your system, fortunately the web site gives you an excellent tutorial to learn how to do this.
Josef W. Zwanziger
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Re: polarization by lda+u and paw pseudopotention
Hello,
Thankyou for your attention,can i ask you tell me the address of tutorial?
Best regards.
Thankyou for your attention,can i ask you tell me the address of tutorial?
Best regards.
-
david.waroquiers
- Posts: 138
- Joined: Sat Aug 15, 2009 12:45 am
Re: polarization by lda+u and paw pseudopotention
Hello,
You can find information about generating PAW Datasets with atomPAW in the following link :
http://www.abinit.org/downloads/PAW/AtomPAW2Abinit-Manual-html/AtomPAW2Abinit2.htm
David
You can find information about generating PAW Datasets with atomPAW in the following link :
http://www.abinit.org/downloads/PAW/AtomPAW2Abinit-Manual-html/AtomPAW2Abinit2.htm
David
Re: polarization by lda+u and paw pseudopotention
Dear David,
Thankyou for your guide but i don't know what i have to change?
could you please tell me which part of paw of Bi and Fe i have to change?
Thank you.
Thankyou for your guide but i don't know what i have to change?
could you please tell me which part of paw of Bi and Fe i have to change?
Thank you.
Re: polarization by lda+u and paw pseudopotention
If the PAW spheres are too big you have to change rpaw, the paw radius. This is set in the input file to atompaw when the paw sets are created. Please, do the tutorial on PAW data sets and then try re-making your PAW sets with smaller radii.
Josef W. Zwanziger
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Re: polarization by lda+u and paw pseudopotention
Dear professor jzwanzig,
as you tell me i have found about make atompaw but i have some question,please:
can i use windose instead of linux to make atompaw?
Is there any new version of abinit that dosen't have this proble?
Best regards.
as you tell me i have found about make atompaw but i have some question,please:
can i use windose instead of linux to make atompaw?
Is there any new version of abinit that dosen't have this proble?
Best regards.
Re: polarization by lda+u and paw pseudopotention
Well, you can compile atompaw on any platform that has a fortran compiler and blas/lapack libraries. Next, this is not an issue of abinit ("a problem" as you call it), the PAW input files are totally separate from abinit and are read in at run-time. If an available set doesn't meet your needs or tests then you have to create your own.
Josef W. Zwanziger
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Re: polarization by lda+u and paw pseudopotention
Hello professor jzwanzig,
Thankyou for your kindness,i couldn't make paw for bi and fe until now but i used paw pseudopotential generated by USpp2abinit.in this pseuodopotentials rpaw is smaller than paw pseuodopotentials but when i run my problem i have WArning and ERROR.
could you please tell me what is this wrong?
Thankyou.Best regards.
input:
acell 3*5.6343 angstrom
angdeg 59.348 59.348 59.348
natom 10
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
ixc 2
iscf 17
nstep 100
ecut 35
pawecutdg 50
pawovlp -1.5
kptopt 1
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-12
berryopt -1
rfdir 1 1 1
ndtset 3
jdtset 1 2 3
xred1 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.220770001 0.220770001 0.220770001
0.720770001 0.720770001 0.720770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred2 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.230770001 0.230770001 0.230770001
0.730770001 0.730770001 0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred3 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
-0.230770001 -0.230770001 -0.230770001
-0.730770001 -0.730770001 -0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.7 0.0 ev
WARNING:
print_ij: WARNING -
The matrix seems to have high value(s) !
( 3 components have a value greater than 50.0).
It can cause instabilities during SCF convergence.
Action: you should check your atomic dataset (psp file)
and look for "high" projector functions...
ERROR:
smatrix_pawinit: ERROR -
Not compatible with mkmem=0 !
Thankyou.
Thankyou for your kindness,i couldn't make paw for bi and fe until now but i used paw pseudopotential generated by USpp2abinit.in this pseuodopotentials rpaw is smaller than paw pseuodopotentials but when i run my problem i have WArning and ERROR.
could you please tell me what is this wrong?
Thankyou.Best regards.
input:
acell 3*5.6343 angstrom
angdeg 59.348 59.348 59.348
natom 10
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
ixc 2
iscf 17
nstep 100
ecut 35
pawecutdg 50
pawovlp -1.5
kptopt 1
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-12
berryopt -1
rfdir 1 1 1
ndtset 3
jdtset 1 2 3
xred1 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.220770001 0.220770001 0.220770001
0.720770001 0.720770001 0.720770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred2 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.230770001 0.230770001 0.230770001
0.730770001 0.730770001 0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred3 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
-0.230770001 -0.230770001 -0.230770001
-0.730770001 -0.730770001 -0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.7 0.0 ev
WARNING:
print_ij: WARNING -
The matrix seems to have high value(s) !
( 3 components have a value greater than 50.0).
It can cause instabilities during SCF convergence.
Action: you should check your atomic dataset (psp file)
and look for "high" projector functions...
ERROR:
smatrix_pawinit: ERROR -
Not compatible with mkmem=0 !
Thankyou.
Re: polarization by lda+u and paw pseudopotention
Hi,
I got your input to run without much problem. The WARNING is due to the fact that the Bi PAW dataset must have projectors close together or something like that; it's just a warning, so you can live with it, but it is also a signal that the Bi PAW dataset isn't very good and you should make a better one.
The mkmem issue I think is due to your machine having limited RAM and so abinit is trying to write to disk but the code is not supporting that operation. Your calculation is quite large (many bands, atoms) and you are using a VERY high cut-off for PAW. If you make better PAW datasets you should be able to get this under 20 or even under 15 (though I find this difficult with oxygen) and that will help a lot, by reducing memory needs substantially.
So, to summarize: the calculation is working but you really need to make and optimize your own PAW sets and you probably need to run on a larger computer with more memory.
I got your input to run without much problem. The WARNING is due to the fact that the Bi PAW dataset must have projectors close together or something like that; it's just a warning, so you can live with it, but it is also a signal that the Bi PAW dataset isn't very good and you should make a better one.
The mkmem issue I think is due to your machine having limited RAM and so abinit is trying to write to disk but the code is not supporting that operation. Your calculation is quite large (many bands, atoms) and you are using a VERY high cut-off for PAW. If you make better PAW datasets you should be able to get this under 20 or even under 15 (though I find this difficult with oxygen) and that will help a lot, by reducing memory needs substantially.
So, to summarize: the calculation is working but you really need to make and optimize your own PAW sets and you probably need to run on a larger computer with more memory.
Josef W. Zwanziger
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Re: polarization by lda+u and paw pseudopotention
Hello professor jzwanzig,
Thank you for your kindness,as you told me i have to make pusedopotential for Bi and Fe myself.i studied tutorial and user guide and i download atompaw-3.0 version
but i have a problem:when i do command ./configure to compile it i have an error like this:
configure: error: no working linear algebra libraries have been found
i underestood i need lapack and blas so i download lapack-3.3.0.but i have again that error.
could you please tell me where i have to downlod lapack-3.3.0?in atompaw-3.0?
could you plaese guide me what i have to do to compile atompaw-3.0?
this is my conatins of atompaw-3.0:
aclocal.m4 ChangeLog config.h.in configure COPYING example Makefile.am NEWS src
AUTHORS config config.log configure.ac doc INSTALL Makefile.in README
Best wishes.
Thank you for your kindness,as you told me i have to make pusedopotential for Bi and Fe myself.i studied tutorial and user guide and i download atompaw-3.0 version
but i have a problem:when i do command ./configure to compile it i have an error like this:
configure: error: no working linear algebra libraries have been found
i underestood i need lapack and blas so i download lapack-3.3.0.but i have again that error.
could you please tell me where i have to downlod lapack-3.3.0?in atompaw-3.0?
could you plaese guide me what i have to do to compile atompaw-3.0?
this is my conatins of atompaw-3.0:
aclocal.m4 ChangeLog config.h.in configure COPYING example Makefile.am NEWS src
AUTHORS config config.log configure.ac doc INSTALL Makefile.in README
Best wishes.
Re: polarization by lda+u and paw pseudopotention
Hi,
it sounds like your atompaw ./configure can't find the lapack libraries. You may have to tell it where they are. Suppose they are in /usr/local/lib, then you would use
./configure --with-linalg-libs="-L/usr/local/lib -llapack -lblas".
The "-L" is the typical compiler option that that tells it to include a given directory in its search for libraries.
what computer platform are you using, and what compiler?
Joe
it sounds like your atompaw ./configure can't find the lapack libraries. You may have to tell it where they are. Suppose they are in /usr/local/lib, then you would use
./configure --with-linalg-libs="-L/usr/local/lib -llapack -lblas".
The "-L" is the typical compiler option that that tells it to include a given directory in its search for libraries.
what computer platform are you using, and what compiler?
Joe
Josef W. Zwanziger
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
- Alain_Jacques
- Posts: 279
- Joined: Sat Aug 15, 2009 9:34 pm
- Location: Université catholique de Louvain - Belgium
Re: polarization by lda+u and paw pseudopotention
... and, if you compile atompaw on the same platform as abinit, you have a ready-to-use blas/lapack (and well tested) in prereqs/linalg.
Alain
Alain
Re: polarization by lda+u and paw pseudopotention
Hello professor Alain-Jacques and professor jzwanzig,
Thankyou for your kindness,i could solve that problem with atompaw,now i can run my job with atompaw but when i wana use it for Bi my job will be exit.
could you please tell me what is my mistake?
this is my Input file:
Bi 83
LDA-PW
6 6 5 4 0
6 1 3
0 0 0
c
c
c
c
c
v
c
c
c
c
v
c
c
v
v
3
1.6
n
n
n
n
Vanderbilt
2.0
1.6
1.6
1.6
1.6
1.6
1.6
0
i have this files when i exit:
Bi Bi.atompaw.input dummy out
Best regards.
Thankyou for your kindness,i could solve that problem with atompaw,now i can run my job with atompaw but when i wana use it for Bi my job will be exit.
could you please tell me what is my mistake?
this is my Input file:
Bi 83
LDA-PW
6 6 5 4 0
6 1 3
0 0 0
c
c
c
c
c
v
c
c
c
c
v
c
c
v
v
3
1.6
n
n
n
n
Vanderbilt
2.0
1.6
1.6
1.6
1.6
1.6
1.6
0
i have this files when i exit:
Bi Bi.atompaw.input dummy out
Best regards.
- Alain_Jacques
- Posts: 279
- Joined: Sat Aug 15, 2009 9:34 pm
- Location: Université catholique de Louvain - Belgium
Re: polarization by lda+u and paw pseudopotention
Hello Mina,
Unless you have compelling reasons to do so, no need to put 4f in valence (and if you want to, 5p should be there too IMHO - look at the energies). Add at least 1 projector/moment if you care about accuracy. So I would suggest to work around the following atompaw input (just a quick draft, very unfinished; you'll have to look at the derivatives to optimize it) ...
Inspire yourself with Joe's good pseudo for Pb on http://www.abinit.org/downloads/PAW/CON ... ATA/082-pb - it should be very similar.
Kind regards,
Alain
Unless you have compelling reasons to do so, no need to put 4f in valence (and if you want to, 5p should be there too IMHO - look at the energies). Add at least 1 projector/moment if you care about accuracy. So I would suggest to work around the following atompaw input (just a quick draft, very unfinished; you'll have to look at the derivatives to optimize it) ...
Code: Select all
Bi 83
LDA-PW loggrid 1200 scalarrelativistic
6 6 5 4 0
6 1 3
0 0 0
c
c
c
c
c
v
c
c
c
c
v
c
c
v
c
2
2.2
y
0.0
n
y
1.0
n
y
0.0
n
VANDERBILT
3 0.0 ultrasoft
2.15
2.15
2.15
2.15
2.15
2.15
0Inspire yourself with Joe's good pseudo for Pb on http://www.abinit.org/downloads/PAW/CON ... ATA/082-pb - it should be very similar.
Kind regards,
Alain
Re: polarization by lda+u and paw pseudopotention
Dear professor Alain-Jacques,
Thankyou for your kindness,as you know i wana to calculate polarization of bifeo3 with LDA+U but fe and bi have large over lapping.i have to make pesudopotential for them and with rpaw smaller than before.i saw you used rpaw=2.2.
can i use this rpaw?or can i choose smaller than this?
Best wishes.
Thankyou for your kindness,as you know i wana to calculate polarization of bifeo3 with LDA+U but fe and bi have large over lapping.i have to make pesudopotential for them and with rpaw smaller than before.i saw you used rpaw=2.2.
can i use this rpaw?or can i choose smaller than this?
Best wishes.
- Alain_Jacques
- Posts: 279
- Joined: Sat Aug 15, 2009 9:34 pm
- Location: Université catholique de Louvain - Belgium
Re: polarization by lda+u and paw pseudopotention
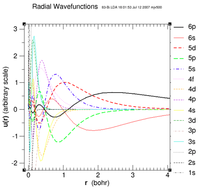
Hello Mina,
rpaw=2.2 a.u. choice is related to core/valence considerations. If you want a rpaw around 1.6 a.u., then you'll have to include a bunch of electrons as valence. Look at the attached plot of Bi radial wf ... At 1.6 a.u., 5s, 5p, 5d, 6s, 6p are all valence. So my second draft for a 60 core / 23 valence Bi would look like
Not a finished job ... it has to be tuned/checked/tested ...
Alain
rpaw=2.2 a.u. choice is related to core/valence considerations. If you want a rpaw around 1.6 a.u., then you'll have to include a bunch of electrons as valence. Look at the attached plot of Bi radial wf ... At 1.6 a.u., 5s, 5p, 5d, 6s, 6p are all valence. So my second draft for a 60 core / 23 valence Bi would look like
Code: Select all
Bi 83
LDA-PW loggrid 1200 scalarrelativistic
6 6 5 4 0
6 1 3
0 0 0
c
c
c
c
v
v
c
c
c
v
v
c
c
v
c
2
1.6
y
2.0
n
y
0.0
n
y
0.0
n
VANDERBILT
3 0.0 ultrasoft
1.5
1.5
1.5
1.5
1.5
1.5
1.5
1.5
0Not a finished job ... it has to be tuned/checked/tested ...
Alain
Re: polarization by lda+u and paw pseudopotention
Hello professor Alain_Jacques,
Thankyou for your kindness,i could make pesudopotential and nowi have pesudopotential for Bi.i wanted to test it for this i wana to calculate bond length of Bi but i have an error.could you please tell me what i have done wrong?
input file:
ndtset 21
xcart: -0.5 0.0 0.0
0.5 0.0 0.0
xcart+ -0.025 0.0 0.0
0.025 0.0 0.0
getwfk -1
nband 50
acell 10 10 10
ntypat 1
znucl 83
natom 2
typat 1 1
ecut 10
pawecutdg 20
kptopt 0
nkpt 1
nstep 10
toldfe 1.0d-6
diemac 2.0
~
ERROR:
- pspatm: opening atomic psp file ../83bi-N/Bi.LDA-PW-paw.abinit
Paw atomic data for element Bi - Generated by atompaw v3.0 & AtomPAW2Abinit v3.3.1
83.00000 23.00000 20110130 znucl, zion, pspdat
7 7 2 0 853 0.00000 pspcod,pspxc,lmax,lloc,mmax,r2well
psp7in: ERROR -
This version of PAW psp file ( 5) is not compatible with
current version of Abinit.
Best regards.
Thankyou for your kindness,i could make pesudopotential and nowi have pesudopotential for Bi.i wanted to test it for this i wana to calculate bond length of Bi but i have an error.could you please tell me what i have done wrong?
input file:
ndtset 21
xcart: -0.5 0.0 0.0
0.5 0.0 0.0
xcart+ -0.025 0.0 0.0
0.025 0.0 0.0
getwfk -1
nband 50
acell 10 10 10
ntypat 1
znucl 83
natom 2
typat 1 1
ecut 10
pawecutdg 20
kptopt 0
nkpt 1
nstep 10
toldfe 1.0d-6
diemac 2.0
~
ERROR:
- pspatm: opening atomic psp file ../83bi-N/Bi.LDA-PW-paw.abinit
Paw atomic data for element Bi - Generated by atompaw v3.0 & AtomPAW2Abinit v3.3.1
83.00000 23.00000 20110130 znucl, zion, pspdat
7 7 2 0 853 0.00000 pspcod,pspxc,lmax,lloc,mmax,r2well
psp7in: ERROR -
This version of PAW psp file ( 5) is not compatible with
current version of Abinit.
Best regards.
Re: polarization by lda+u and paw pseudopotention
What version of abinit are you running?
Josef W. Zwanziger
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Professor, Department of Chemistry
Canada Research Chair in NMR Studies of Materials
Dalhousie University
Halifax, NS B3H 4J3 Canada
jzwanzig@gmail.com
Re: polarization by lda+u and paw pseudopotention
Dear professor jzwanzig,
i have abinit-6.0.4.
Best regards.
i have abinit-6.0.4.
Best regards.
Re: polarization by lda+u and paw pseudopotention
Dear professor jzwanzig,
sorry.i have another question too.as you know i sended input file with this errors and you told me "I got your input to run without much problem. The WARNING is due to the fact that the Bi PAW dataset must have projectors close together or something like that; it's just a warning, so you can live with it, but it is also a signal that the Bi PAW dataset isn't very good and you should make a better one.
The mkmem issue I think is due to your machine having limited RAM and so abinit is trying to write to disk but the code is not supporting that operation. Your calculation is quite large (many bands, atoms) and you are using a VERY high cut-off for PAW. If you make better PAW datasets you should be able to get this under 20 or even under 15 (though I find this difficult with oxygen) and that will help a lot, by reducing memory needs substantially."
could you please tell me how can i optimize my own paw set and reduce memory needs becaese i have not larger computer with large memory?
my input :acell 3*5.6343 angstrom
angdeg 59.348 59.348 59.348
natom 10
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
ixc 2
iscf 17
nstep 100
ecut 35
pawecutdg 50
pawovlp -1.5
kptopt 1
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-12
berryopt -1
rfdir 1 1 1
ndtset 3
jdtset 1 2 3
xred1 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.220770001 0.220770001 0.220770001
0.720770001 0.720770001 0.720770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred2 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.230770001 0.230770001 0.230770001
0.730770001 0.730770001 0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred3 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
-0.230770001 -0.230770001 -0.230770001
-0.730770001 -0.730770001 -0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.7 0.0 ev
WARNING:
print_ij: WARNING -
The matrix seems to have high value(s) !
( 3 components have a value greater than 50.0).
It can cause instabilities during SCF convergence.
Action: you should check your atomic dataset (psp file)
and look for "high" projector functions...
ERROR:
smatrix_pawinit: ERROR -
Not compatible with mkmem=0 !
Thank you for your kindness.
sorry.i have another question too.as you know i sended input file with this errors and you told me "I got your input to run without much problem. The WARNING is due to the fact that the Bi PAW dataset must have projectors close together or something like that; it's just a warning, so you can live with it, but it is also a signal that the Bi PAW dataset isn't very good and you should make a better one.
The mkmem issue I think is due to your machine having limited RAM and so abinit is trying to write to disk but the code is not supporting that operation. Your calculation is quite large (many bands, atoms) and you are using a VERY high cut-off for PAW. If you make better PAW datasets you should be able to get this under 20 or even under 15 (though I find this difficult with oxygen) and that will help a lot, by reducing memory needs substantially."
could you please tell me how can i optimize my own paw set and reduce memory needs becaese i have not larger computer with large memory?
my input :acell 3*5.6343 angstrom
angdeg 59.348 59.348 59.348
natom 10
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
ixc 2
iscf 17
nstep 100
ecut 35
pawecutdg 50
pawovlp -1.5
kptopt 1
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-12
berryopt -1
rfdir 1 1 1
ndtset 3
jdtset 1 2 3
xred1 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.220770001 0.220770001 0.220770001
0.720770001 0.720770001 0.720770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred2 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
0.230770001 0.230770001 0.230770001
0.730770001 0.730770001 0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
xred3 0.000000000 0.000000000 0.000000000
0.500000000 0.500000000 0.500000000
-0.230770001 -0.230770001 -0.230770001
-0.730770001 -0.730770001 -0.730770001
0.527899981 0.933300018 0.394800007
0.394800007 0.527899981 0.933300018
0.933300018 0.394800007 0.527899981
0.894800007 0.433299989 0.027899999
0.433299989 0.027899999 0.894800007
0.027899999 0.894800007 0.433299989
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.7 0.0 ev
WARNING:
print_ij: WARNING -
The matrix seems to have high value(s) !
( 3 components have a value greater than 50.0).
It can cause instabilities during SCF convergence.
Action: you should check your atomic dataset (psp file)
and look for "high" projector functions...
ERROR:
smatrix_pawinit: ERROR -
Not compatible with mkmem=0 !
Thank you for your kindness.
Re: polarization by lda+u and paw pseudopotention
Hello professor Alain and Jzwanzig,
i could make pseudopotential for Bi with rpaw 1.6.i wantde to test that pseuodopotential so i calculated bond length of Bi and i had overlapping again and i used variable pawovlp=-1.5 so i got it 2.77 bohr but i think it is smaller than real value.could you please tell me this means that pseuodopotential is wrong?
Best regards.
i could make pseudopotential for Bi with rpaw 1.6.i wantde to test that pseuodopotential so i calculated bond length of Bi and i had overlapping again and i used variable pawovlp=-1.5 so i got it 2.77 bohr but i think it is smaller than real value.could you please tell me this means that pseuodopotential is wrong?
Best regards.
- Alain_Jacques
- Posts: 279
- Joined: Sat Aug 15, 2009 9:34 pm
- Location: Université catholique de Louvain - Belgium
Re: polarization by lda+u and paw pseudopotention
Hello Mina,
I have some difficulties to follow what failed - why you still have overlapping with a rpaw=1.6 a.u. Are you using the input reproduced on 31/01 9:49am?
May I suggest first to do a plain ground state check of the quality of your new pseudo - a simple bulk Bi test input based on Wyckoff CIF - http://www.crystallography.net/cif/9/9008576.cif data with a variable acell and ample ecut; plot acell vs. energy and see you can reproduce the expected value. If I remember well, shortest Bi-Bi distance is around 5.8 a.u - no way to have spheres overlapping in these conditions with such a small rpaw. And if it is sound, use a similar input with the optimal acell and vary ecut; follow output energy vs. ecut and find the ecut corresponding to an energy variation below 1mHa - this parameter will be very useful to optimize further calculations with your new pseudo.
Kind regards,
Alain
I have some difficulties to follow what failed - why you still have overlapping with a rpaw=1.6 a.u. Are you using the input reproduced on 31/01 9:49am?
May I suggest first to do a plain ground state check of the quality of your new pseudo - a simple bulk Bi test input based on Wyckoff CIF - http://www.crystallography.net/cif/9/9008576.cif data with a variable acell and ample ecut; plot acell vs. energy and see you can reproduce the expected value. If I remember well, shortest Bi-Bi distance is around 5.8 a.u - no way to have spheres overlapping in these conditions with such a small rpaw. And if it is sound, use a similar input with the optimal acell and vary ecut; follow output energy vs. ecut and find the ecut corresponding to an energy variation below 1mHa - this parameter will be very useful to optimize further calculations with your new pseudo.
Kind regards,
Alain
Re: polarization by lda+u and paw pseudopotention
Dear professor Alain,
i used this input file to make pseudopotential.and could you plaese tell me you means is that i use ground state program to find best ecut?
my input:
Bi 83
LDA-PW loggrid 1200 scalarrelativistic
6 6 5 4 0
6 1 3
0 0 0
c
c
c
c
v
v
c
c
c
v
v
c
c
v
c
2
1.6
y
2.0
n
y
0.0
n
y
0.0
n
Vanderbilt
3 0.0 ultrasoft
1.5
1.5
1.5
1.5
1.5
1.5
1.5
1.5
2
default
3
UPFDX 0.005d0 UPFXMIN -9.d0 UPFZMESH 1.d0
0
Thankyou for your kindness.
i used this input file to make pseudopotential.and could you plaese tell me you means is that i use ground state program to find best ecut?
my input:
Bi 83
LDA-PW loggrid 1200 scalarrelativistic
6 6 5 4 0
6 1 3
0 0 0
c
c
c
c
v
v
c
c
c
v
v
c
c
v
c
2
1.6
y
2.0
n
y
0.0
n
y
0.0
n
Vanderbilt
3 0.0 ultrasoft
1.5
1.5
1.5
1.5
1.5
1.5
1.5
1.5
2
default
3
UPFDX 0.005d0 UPFXMIN -9.d0 UPFZMESH 1.d0
0
Thankyou for your kindness.
Re: polarization by lda+u and paw pseudopotention
Hello professor,
when i wana to calculate polarization of bifeo3 since i have 10 atom it needs big memory so i used variable spgroup and natrd but when i used natrd 3.i have an error that according to symmetry you need 14 atom and as you know bifeo3 have 10 atoms in its primitive unit cell.when i changed natrd to 14 again i have another error.
could you please look at my input and tell me what i have done wrong?
input file:
acell 3*5.6343 angstrom
angdeg 60 60 60
spgroup 161
natom 14
brvltt -1
natrd 3
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
ixc 7
iscf 17
nstep 90
ecut 20
pawecutdg 35
pawovlp -1.5
kptopt 1
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-10
berryopt -1
rfdir 1 1 1
ndtset 3
jdtset 1 2 3
xred1 0.000000000 0.000000000 0.000000000
0.220770001 0.220770001 0.220770001
0.527899981 0.933300018 0.394800007
xred2 0.000000000 0.000000000 0.000000000
0.230770001 0.230770001 0.230770001
0.527899981 0.933300018 0.394800007
xred3 0.000000000 0.000000000 0.000000000
-0.230770001 -0.230770001 -0.230770001
0.527899981 0.933300018 0.394800007
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.07 0.0 ev
and error:
chkorthsy: ERROR -
The symmetry operation number 2 does not preserve
vector lengths and angles.
The value of the residual is 3.0622E+04.
Action : modify rprim, acell and/or symrel so that
vector lengths and angles are preserved.
Beware, the tolerance on symmetry operations is very small.
Thankyou for your kindness.
when i wana to calculate polarization of bifeo3 since i have 10 atom it needs big memory so i used variable spgroup and natrd but when i used natrd 3.i have an error that according to symmetry you need 14 atom and as you know bifeo3 have 10 atoms in its primitive unit cell.when i changed natrd to 14 again i have another error.
could you please look at my input and tell me what i have done wrong?
input file:
acell 3*5.6343 angstrom
angdeg 60 60 60
spgroup 161
natom 14
brvltt -1
natrd 3
ntypat 3
znucl 83 26 8
typat 1 1 2 2 3 3 3 3 3 3
ixc 7
iscf 17
nstep 90
ecut 20
pawecutdg 35
pawovlp -1.5
kptopt 1
ngkpt 2 2 2
nshiftk 1
shiftk 0.5 0.5 0.5
toldfe 1.0d-10
berryopt -1
rfdir 1 1 1
ndtset 3
jdtset 1 2 3
xred1 0.000000000 0.000000000 0.000000000
0.220770001 0.220770001 0.220770001
0.527899981 0.933300018 0.394800007
xred2 0.000000000 0.000000000 0.000000000
0.230770001 0.230770001 0.230770001
0.527899981 0.933300018 0.394800007
xred3 0.000000000 0.000000000 0.000000000
-0.230770001 -0.230770001 -0.230770001
0.527899981 0.933300018 0.394800007
nbdbuf 0
usepawu 1
lpawu -1 2 -1
upawu 0.0 7.0 0.0 ev
jpawu 0.0 0.07 0.0 ev
and error:
chkorthsy: ERROR -
The symmetry operation number 2 does not preserve
vector lengths and angles.
The value of the residual is 3.0622E+04.
Action : modify rprim, acell and/or symrel so that
vector lengths and angles are preserved.
Beware, the tolerance on symmetry operations is very small.
Thankyou for your kindness.