Page 1 of 1
[SOLVED] paw dataset creation error with atompaw
Posted: Tue Sep 11, 2012 12:48 pm
by ilukacevic
Dear all,
I'm trying to create paw datasets for Ir and O. O dataset creation went well, however, I have a problem with Ir dataset. When I run atompaw, I get an error:
To generate the local pseudopotential, this code can use:
1- a Troullier-Martins scheme for specified l value and energy
2- a non norm-conserving pseudopotential scheme for specified l value and energy
3- a simple pseudization scheme using a single spherical Bessel function
For choice 1, enter (high) l value and energy e
For choice 2, enter (high) l value, energy e and "ultrasoft"
For choice 3, enter "bessel"
At line 407 of file atompaw_prog.f90
Fortran runtime error: Bad integer for item 1 in list input
Here's my atompaw input file for Ir:
Ir 77
GGA-PBE loggrid 2001
6 5 5 4 0 0
5 2 7
0 0 0
c
c
c
c
c
c
c
c
c
c
c
c
v
v
2
2.5164685227
y
2
n
y
2
n
VANDERBILT
2 0
2.5164685227
2.5164685227
2.5164685227
2.5164685227
2
default
0
Does anyone know what this error means and what is the problem here? I use atompaw compiled within abinit 6.10.3.
Thanks in advance.
Yours,
Igor L.
Re: paw dataset creation error with atompaw
Posted: Tue Sep 11, 2012 5:54 pm
by jzwanzig
From your input file your valence space appears to be 5d7 4f14, but the 2 after the last "v" indicates that the highest state to pseudize (lmax) is 2, I think this should be 3.
Re: paw dataset creation error with atompaw
Posted: Wed Sep 12, 2012 12:48 pm
by ilukacevic
Hmmm, now I see it...I made a mistake. I attributed wrong levels as valence ones. It should be
c
c
c
c
c
v
c
c
c
c
c
c
v
c
so that 5d7 6s2 are valence ones. But then the number 2 after the last "v" is correct, right? Yet, again, the code stops with the same message (after I corrected the above mistake). Do you have an idea why?
Igor
Re: paw dataset creation error with atompaw
Posted: Sat Sep 15, 2012 12:06 pm
by Alain_Jacques
Hi Igor,
No, you want to build a dataset up to f electrons so the maximum quantum number for the partial waves in the PAW basis should be 3, not 2. And similarly, the reference quantum number for the local pseudopotential scheme (l loc in the documentation) should be the maximum +1, so I suggest to change the line after the Vanderbilt scheme keyword to 4 0. (I would also switch the scalar relativistic approximation on, use a smaller grid and more projectors).
Kind regards,
Alain
Re: paw dataset creation error with atompaw
Posted: Sat Sep 15, 2012 7:21 pm
by Alain_Jacques
Hi again ...
IMHO the PAW radius could be reduced - Iridium is quite small. Valence electrons are 5d7 6s2 so I would start, for a 9 electrons PAW pseudo, with something like:
Ir 77
GGA-PBE scalarrelativistic loggrid 801
6 5 5 4 0
5 2 7
6 0 2
0 0 0
c
c
c
c
c
v
c
c
c
c
c
c
v
c
3
2.2
y
1.0
n
y
1.0
y
2.0
n
y
1.0
n
y
1.0
y
2.0
n
Vanderbilt
4 0 ultrasoft
2.2
2.2
2.2
2.2
2.2
2.2
2.2
2.2
2
default
0
only a suggestion template with 2 projectors per state - I didn't even try to adjust the reference energies or the radii.
Kind regards,
Alain
Re: paw dataset creation error with atompaw
Posted: Mon Sep 17, 2012 10:46 am
by ilukacevic
Hi, Alain!
Thanks for the effort. It truly helps. I have some questions, however, regarding your suggestion:
1] Is there any special reason to why did you add 6s shell (line 5), while it's a full shell and not an empty or a partially occupied one? Marcs' instructions say that full shells can be omitted ("but do not have to be"). Therefore, I suppose your line won't hurt.
2] Why do you suggest ultrasoft scheme for obtaining the V_loc? I built my oxygen PAW with troullier-martens scheme. Won't they interfere if they're built with different schemes?
Best regards,
Igor
Re: paw dataset creation error with atompaw
Posted: Tue Sep 18, 2012 8:03 pm
by Alain_Jacques
Hello Igor,
[1] ... I added 6s2 for truly cosmetic reasons. It allows me to know that it is a 9 valence electrons pseudo without having to decode the c c c v etc. spaghetti . Quite useful when you'll have several "plain" and semi core pseudos of the same atom with different core/valence configurations. Both ways are equivalent, the valence states will be included in the pseudo basis.
[2] ... it's only a suggestion

When I start a new pseudo, in the process of optimizing the different variables, I generally start with Vanderbilt/ultrasoft combo because I feel that atompaw converges and often gives decent solutions (i.e. no crazy amplitudes, later on already good ecut, ...). Then I play with the partial waves reference energies to cover a good range (gs or GW?) and to return good looking logderivs, ...
There is no problem mixing pseudos made with different pseudization schemes - but it's not advisable to mix different exchange - correlation functionals - Abinit should complain in this case.
Kind regards,
Alain
Re: paw dataset creation error with atompaw
Posted: Wed Sep 19, 2012 9:22 am
by ilukacevic
Alain_Jacques wrote:Hi Igor,
No, you want to build a dataset up to f electrons so the maximum quantum number for the partial waves in the PAW basis should be 3, not 2.
You're right, I made another mistake here.
Alain_Jacques wrote:And similarly, the reference quantum number for the local pseudopotential scheme (l loc in the documentation) should be the maximum +1, so I suggest to change the line after the Vanderbilt scheme keyword to 4 0. (I would also switch the scalar relativistic approximation on, use a smaller grid and more projectors).
Kind regards,
Alain
Got it. I followed the Marc's document for atompaw and there it states l_loc = l quantum number and nothing else. But in abinit tutorial PAW2 is states l_loc=l_max+1.
Now that you helped me solve the problem and teach me a couple of things more, I can turn to paw testing.
Thanks again.
Cheers!
Igor
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Wed Sep 19, 2012 4:36 pm
by Alain_Jacques
Hi Igor,
If you have some time, maybe you'll want to compare the results of your pseudo with the one I'm working on ... 5p6 5d7 6s2 valence i.e. 62/15 e- Iridium. Work in progress, no warranty ... I decline any responsibility for physical or moral damages

More seriously it should be first tested on a plain Ir bulk. ecut= 16Ha @1mHa
Ir 77
GGA-PBE scalarrelativistic loggrid 801
6 6 5 4 0
6 0 2
6 1 0
5 1 6
5 2 7
0 0 0
c
c
c
c
c
v
c
c
c
v
v
c
c
v
c
3
2.42
y
1.7
n
y
3.8
n
y
0.0
n
y
0.0
y
3.2
n
Vanderbilt
4 0 ultrasoft
2.40
2.40
2.40
2.40
2.40
2.40
2.40
2.40
2.40
2
default
0
Enjoy,
Alain
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Thu Sep 20, 2012 8:52 am
by ilukacevic
Hi, Alain!
I will have some time, so I will try it later on. But I also have a question: why do you include 5p6 electron as valence ones? Aren't they lower in energy than 4f14 ones?
Igor
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Thu Sep 20, 2012 7:38 pm
by Alain_Jacques
Hi Igor,
Iridium - as most d-elements - starts to be tricky. Generally, I treat the semicore p of the 4d transition elements as valence states when its eigenenergy lies above 1.5 Ha. I agree that the case of 5d elements may be quite different because the 5p states are strongly localized as the 4f shell becomes filled. And so for the Iridium: most of the published pseudos have 9 valence electrons - but not all, I've seen already 15 valence electrons pseudos. So I wanted to try and compare. Well, it's probably not completely bogus as this 5p valence pseudo gives me an equilibrium fcc lattice constant of 3.879 Angstrom to be compared to the experimental 3.839 Angstrom from Wickoff and the GGA 3.895 Angstrom mentioned in F. Tran PhysRevB.75.115131 (see attached image).
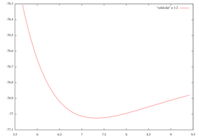
- total energy (in Ha) vs. cell size (acell in Bohr) for fcc Ir bulk
Kind regards,
Alain
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Fri Sep 21, 2012 8:25 am
by ilukacevic
Hi, Alain!
That's very close to experiment. What's the agreement for the 9-valence electron bulk Ir?
Since I'm interested in Ir compounds, I wonder what would happen with the localization of 5p states when Ir starts to bond with some other element, like O (which is more electronegative than Ir). It will be worthwhile to test, as you are doing. Currently I'm trying to make projectors to be of the same order of magnitude as partials waves. For example, for s states I have to set very high E_ref (above 10).
Thanks again!
Best regards,
Igor
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Fri Sep 21, 2012 10:31 am
by Alain_Jacques
my 9e- valence Ir PAW pseudo still has ugly p components. I have to improve it before doing a decent comparison. I'll keep you informed.
Kind regards,
Alain
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Sat Sep 22, 2012 7:58 pm
by Alain_Jacques
Hi Igor,
my current 9e- valence Iridium PAW pseudo ...
Ir 77
GGA-PBE scalarrelativistic loggrid 801
6 6 5 4 0
6 0 2
6 1 0
5 2 7
0 0 0
c
c
c
c
c
v
c
c
c
c
v
c
c
v
c
3
2.50
y
1.3
n
y
3.3
n
y
1.0
n
y
0.0
y
3.0
n
Vanderbilt
4 0 ultrasoft
2.50
2.50
2.50
2.50
2.50
2.50
2.50
2.50
2
default
0
... it gives an equilibrium fcc Ir lattice constant of 3.8599 Angstrom. I'm still not fully satisfied - there is a ghost state in p states.
Kind regards,
Alain
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Fri Sep 28, 2012 9:26 am
by ilukacevic
Hi, Alain!
Smth weird happened...a few days ago I posted you a reply, but not until today I saw that there is no reply here at the forum. I'm posting it again.
I have 2 questions:
1] By looking at your 9-electron configuration, it seems that you set 3d10 4f14 6s2 electron as valence ones, am I right?
c 1s2
c 2s2
c 3s2
c 4s2
c 5s2
v 6s2
c 2p6
c 3p6
c 4p6
c 5p6
v 3d10
c 4d10
c 5d7
v 4f14
c ???
This does not coincide even with your 15-electron Ir.
2] In my 9-electron configuration I have 8 partial waves. However, when I run atompaw, I get 13 wavefunctions. How can I know which wavefunction corresponds to which partial wave?
Cheers!
Igor
Re: [SOLVED] paw dataset creation error with atompaw
Posted: Sat Sep 29, 2012 12:38 pm
by Alain_Jacques
Hi Igor,
Yeah, I only saw your questions today.
1] By looking at your 9-electron configuration, it seems that you set 3d10 4f14 6s2 electron as valence ones, am I right?
No ... if you look at the fifth line in my pseudo input file, you'll notice that I added an extra empty 6p valence state to build the basis. And so
c = 1s2
c = 2s2
c = 3s2
c = 4s2
c = 5s2
v = 6s2
c = 2p6
c = 3p6
c = 4p6
c = 5p6
v = 6p0
c = 3d10
c = 4d10
v = 5d7
c = 4f14
i.e. 5d7 6s2 6p0 = 9 valence electrons
2]How can I know which wavefunction corresponds to which partial wave?
I agree that the wfn1, wfn2, wfn3, ... naming scheme is all but self explanatory.
The basis functions are listed in the order of the outer to inner principal numbers first i.e. start with n=6 then 5, ... For a given principal quantum number, the valence bound states are listed first then any additional unbound (*) partial waves. To build my pseudo, I've added one s, one p, one d and two f partial waves to the basis, so the order of the 8 basis functions is 6s2 / s* / 6p0 / p* / 5d7 / d* / f1* / f2*
Hope this helps,
Alain
