Problem with ABINITBandStructureMaker
Moderators: MMNSchmitt, gonze
Problem with ABINITBandStructureMaker
I have successfully installed ABINIT 7.4.3 from http://forum.abinit.org/viewtopic.php?f=2&t=1619, and run the example t30 file and got a correct t30.out. I then installed Python and numpy (using Enthought Canopy). I downloaded ABINITBandStructureMaker, ran it and successfully got the .dbs file. Now I run it again hoping to produce the .agr file, but got the following error:
RuntimeWarning: invalid value encountered in double_scalars
arg = dot(vector1, vector2)/norm(vector1)/norm(vector2)
TraceBack (most recent call last):
File: ABINITBandStructureMaker.py, line 1290, in <module>
minLUMOList.append(ctrl.Y[i][ctrl.nvalenceband]>
IndexError: index out of bounds
What is the solution to this problem? It seems to be related to the number of valence bands, which is 4.
RuntimeWarning: invalid value encountered in double_scalars
arg = dot(vector1, vector2)/norm(vector1)/norm(vector2)
TraceBack (most recent call last):
File: ABINITBandStructureMaker.py, line 1290, in <module>
minLUMOList.append(ctrl.Y[i][ctrl.nvalenceband]>
IndexError: index out of bounds
What is the solution to this problem? It seems to be related to the number of valence bands, which is 4.
Re: Problem with ABINITBandStructureMaker
I have the similar difficulty with AbinitBandStructureMaker.py at the second step of its running. The first step with the creation .dbs file was made smoothly (probably with "nband" parameter, though in my .dbs file it was figured as 24?):
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
Traceback (most recent call last):
File "/home/letuan/Tools/Abinit_postproc/AbinitBandStructureMaker.py", line 1291, in <module>
maxHOMO = max(maxHOMOlist)
ValueError: max() arg is an empty sequence
I use Python 2.6 as default version in Rocks 6.2 and Abinit v. 7.10.4.
I appreciate any suggestion for overcoming it.
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
Traceback (most recent call last):
File "/home/letuan/Tools/Abinit_postproc/AbinitBandStructureMaker.py", line 1291, in <module>
maxHOMO = max(maxHOMOlist)
ValueError: max() arg is an empty sequence
I use Python 2.6 as default version in Rocks 6.2 and Abinit v. 7.10.4.
I appreciate any suggestion for overcoming it.
Re: Problem with ABINITBandStructureMaker
I've got another issue: ABINITBandStructureMaker can't parse the input file with 2 datasets.
traceback
variable starter[i] actualy is, so .split()[1] is
the input
traceback
Code: Select all
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
2 dataset(s) detected
Which dataset(s) do you want to use (1 to 2) ? 2
Traceback (most recent call last):
File "AbinitBandStructureMaker.py", line 836, in <module>
datasetkey[-1].append(float(starter[i].split()[1]))
ValueError: could not convert string to float: =variable starter[i] actualy is
Code: Select all
natom = 8 nloc_mem = 1 nspden = 1 nspinor = 1Code: Select all
=the input
- Attachments
-
 AgN3.out
AgN3.out- input file
- (73.62 KiB) Downloaded 516 times
-
Mohamedy002
- Posts: 9
- Joined: Wed Aug 10, 2016 10:56 am
Re: Problem with ABINITBandStructureMaker
Please read the pdf in the following link.
http://www.gnu-darwin.org/www001/ports-1.5a-CURRENT/science/abinit/work/abinit-5.2.4/doc/users/AbinitBandStructureMaker_manual.pdf
I think you need to edit the .dbs file
http://www.gnu-darwin.org/www001/ports-1.5a-CURRENT/science/abinit/work/abinit-5.2.4/doc/users/AbinitBandStructureMaker_manual.pdf
I think you need to edit the .dbs file
-
Mohamedy002
- Posts: 9
- Joined: Wed Aug 10, 2016 10:56 am
 Re: Problem with ABINITBandStructureMaker
Re: Problem with ABINITBandStructureMaker
Hi Vladimir,
Actually to fix this error here is a simple trick that ... I suggest modifying the "AbinitBandStructureMaker.py" file by removing the float before the starter. It works well with me

for the .dbs file
$ python AbinitBandStructureMaker.py
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
Enter the filename :
AgN3.out
2 dataset(s) detected
Which dataset(s) do you want to use (1 to 2) ? 2
"AgN3.out.dbs" file created successfully
for the .agr file
$ python AbinitBandStructureMaker.py AgN3.out.dbs
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
"AgN3.out.agr" file created successfully
Then by using the xmgrace plotting tool you can execute the following command
xmgrace AgN3.out.agr
Finally you got the attached band structure
Mohamed Yousef Hassan,
EEE department,
Nanyang Technological University,
Singapore.
Actually to fix this error here is a simple trick that ... I suggest modifying the "AbinitBandStructureMaker.py" file by removing the float before the starter. It works well with me
for the .dbs file
$ python AbinitBandStructureMaker.py
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
Enter the filename :
AgN3.out
2 dataset(s) detected
Which dataset(s) do you want to use (1 to 2) ? 2
"AgN3.out.dbs" file created successfully
for the .agr file
$ python AbinitBandStructureMaker.py AgN3.out.dbs
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
"AgN3.out.agr" file created successfully
Then by using the xmgrace plotting tool you can execute the following command
xmgrace AgN3.out.agr
Finally you got the attached band structure
Mohamed Yousef Hassan,
EEE department,
Nanyang Technological University,
Singapore.
- Attachments
-
 AgN3.out.in
AgN3.out.in- This is the .dbs file generated by Python I replaced the extension by .in in order to upload it here ... enjoy !
- (11.36 KiB) Downloaded 486 times
-
 AgN3.out.in
AgN3.out.in- This is the .agr file generated by Python I replaced the extension by .in in order to upload it here ... enjoy !
- (32.5 KiB) Downloaded 479 times
-
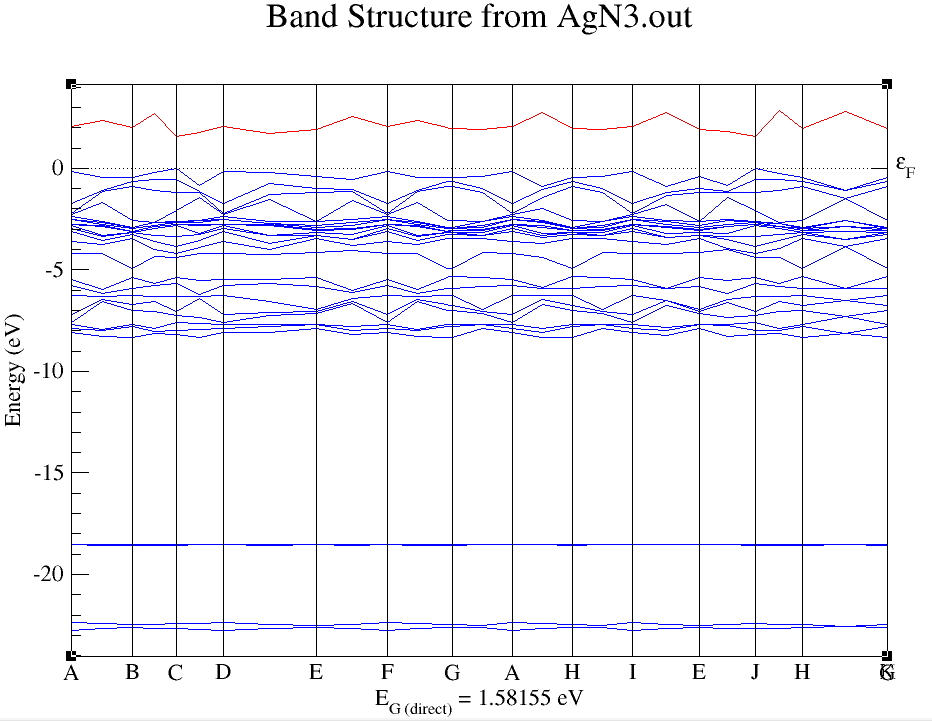
- AgN3.png (31.96 KiB) Viewed 16148 times
Re: Problem with ABINITBandStructureMaker
Mohamedy002 wrote:Hi Vladimir,
Actually to fix this error here is a simple trick that ... I suggest modifying the "AbinitBandStructureMaker.py" file by removing the float before the starter. It works well with me
Mohamed Yousef Hassan,
EEE department,
Nanyang Technological University,
Singapore.
Hi Mohamed.
It's very strange workaround because starter[i] is "natom = 8 nloc_mem = 1 nspden = 1 nspinor = 1" but we want natom value, so starter[i].split()[2] is the answer.
this string came from:
DATASET 1 : space group Ib a m (# 72); Bravais oI (body-center ortho.)
================================================================================
Values of the parameters that define the memory need for DATASET 1.
intxc = 0 ionmov = 0 iscf = 7 lmnmax = 4
lnmax = 4 mgfft = 60 mpssoang = 4 mqgrid = 3001
natom = 8 nloc_mem = 1 nspden = 1 nspinor = 1
nsppol = 1 nsym = 8 n1xccc = 0 ntypat = 2
occopt = 1 xclevel = 1
- mband = 27 mffmem = 1 mkmem = 1
mpw = 4056 nfft = 216000 nkpt = 1
================================================================================
P This job should need less than 62.096 Mbytes of memory.
Rough estimation (10% accuracy) of disk space for files :
_ WF disk file : 1.673 Mbytes ; DEN or POT disk file : 1.650 Mbytes.
================================================================================
DATASET 2 : space group Ib a m (# 72); Bravais oI (body-center ortho.)
================================================================================
Values of the parameters that define the memory need for DATASET 2.
intxc = 0 ionmov = 0 iscf = -2 lmnmax = 4
lnmax = 4 mgfft = 60 mpssoang = 4 mqgrid = 3001
natom = 8 nloc_mem = 1 nspden = 1 nspinor = 1
nsppol = 1 nsym = 8 n1xccc = 0 ntypat = 2
occopt = 1 xclevel = 1
- mband = 27 mffmem = 1 mkmem = 21
mpw = 8200 nfft = 216000 nkpt = 21
================================================================================
but there is another natom definition
--------------------------------------------------------------------------------
------------- Echo of variables that govern the present computation ------------
--------------------------------------------------------------------------------
....
kptopt1 1
kptopt2 -1
kptrlatt 1 0 0 0 1 0 0 0 1
kptrlen1 9.59379937E+00
kptrlen2 3.00000000E+01
P mkmem1 1
P mkmem2 21
natom 8
nband1 27
nband2 27
nbdbuf1 0
nbdbuf2 2
ndtset 2
ngfft 60 60 60
nkpt1 1
nkpt2 21
nsym 8
ntypat 2
I think if we want to get proper value of input variables we should parse only "Echo of variables that govern the present computation" section or DATASET# section not both.
-
Mohamedy002
- Posts: 9
- Joined: Wed Aug 10, 2016 10:56 am
Re: Problem with ABINITBandStructureMaker
Hi Vladimir,
First, since the python script can detect the data sets in the output files and ask you to enter the number of the data set you want to manipulate, then the parser will go inside the given data set only and will not go through the " Echo of variables that govern the present computation". Therefore, I think this is not the problem.
Second, from my point of view python see the values inside the output data files as string not integers or float. Therefore when you put float(starter[i],split()[1]) it gives you error
Mohamed Yousef Hassan
EEE, Nanyang Technological University,
Singapore.
First, since the python script can detect the data sets in the output files and ask you to enter the number of the data set you want to manipulate, then the parser will go inside the given data set only and will not go through the " Echo of variables that govern the present computation". Therefore, I think this is not the problem.
Second, from my point of view python see the values inside the output data files as string not integers or float. Therefore when you put float(starter[i],split()[1]) it gives you error
Mohamed Yousef Hassan
EEE, Nanyang Technological University,
Singapore.
Re: Problem with ABINITBandStructureMaker
Hi Mohamed.
look in the end of your AgN3.out.in file attached (11.36 KiB).
natom: = = 8
this means that 1st and 2nd DATASET parsed with result '=' (this cannot convert to float) and 'Echo of variables that govern the present computation' section parsed with result '8'.
starter is a list of lines from file start to 188 line ("== DATASET 1 ==================================================================")
look in the end of your AgN3.out.in file attached (11.36 KiB).
DATABASE KEY:
ecut: 40.0
natom: = = 8
nband: 27.0
occopt: = =
toldfe: 0.0
toldff: 0.0
tolvrs: 0.0
tolwfr: 1e-12
typat: 2 2 1 1 1 1 1 1
reciprocalvectors: 0.0 0.0884912 0.0882256 0.0944959 0.0 0.0882256 0.0944959 0.0884912 0.0
reducedcoordinates: 0.25 0.25 0.0 0.75 0.75 -0.0 0.1327 0.3422 0.4749 0.0 0.5 0.5 -0.1327 -0.3422 -0.4749 0.6327 0.1578 0.7905 0.5 0.0 0.5 -0.6327 -0.1578 -0.7905
natom: = = 8
this means that 1st and 2nd DATASET parsed with result '=' (this cannot convert to float) and 'Echo of variables that govern the present computation' section parsed with result '8'.
Code: Select all
starter = ctrl.filedata[:ctrl.datasetlocation[0][2]-1]starter is a list of lines from file start to 188 line ("== DATASET 1 ==================================================================")
-
Mohamedy002
- Posts: 9
- Joined: Wed Aug 10, 2016 10:56 am
Re: Problem with ABINITBandStructureMaker
I see I can figure now why we have two equal sign in front of each variable like "natom". But I think after getting the .dbs you can edit it. Right?
so at least we have the .dbs file which we can then edit further.
That's why we don't have any error concerning "ecut" and "nband" since they are mentioned only in the "Echo of variables that govern the present computation" only in this section compared to "natom" which is mentioned in Data set 1 section, Data set 2 section and the "Echo of variables that govern the present computation" section.
Also please find the attached reference band structure of silver azide from literature.
Mohamed Yousef Hassan,
EEE Nanyang Technological University,
Singapore.
so at least we have the .dbs file which we can then edit further.
That's why we don't have any error concerning "ecut" and "nband" since they are mentioned only in the "Echo of variables that govern the present computation" only in this section compared to "natom" which is mentioned in Data set 1 section, Data set 2 section and the "Echo of variables that govern the present computation" section.
Also please find the attached reference band structure of silver azide from literature.
Mohamed Yousef Hassan,
EEE Nanyang Technological University,
Singapore.
- Attachments
-
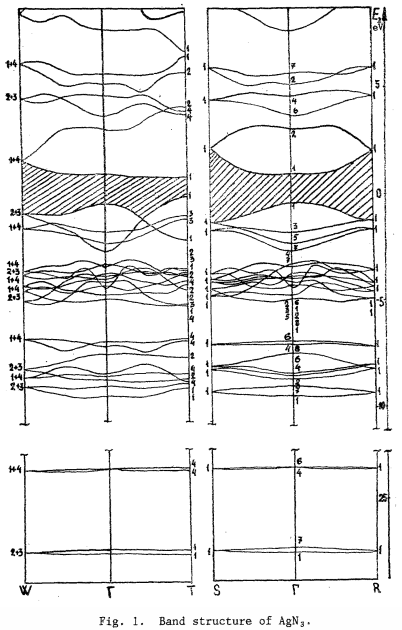
- AgN3.png (110.75 KiB) Viewed 16116 times
Re: Problem with ABINITBandStructureMaker
Hi Mohamed and Vladimir
For a newbies, how can I fix it ?
I ran abinitbandstructuremaker.py and got the error see below
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
2 dataset(s) detected
Which dataset(s) do you want to use (1 to 2) ? 2
Traceback (most recent call last):
File "../band.py", line 836, in <module>
datasetkey[-1].append(float(starter[i].split()[1]))
ValueError: could not convert string to float: =
How can I fix it by step by step ?
Mutta
Physics Thailand
For a newbies, how can I fix it ?
I ran abinitbandstructuremaker.py and got the error see below
=========================================
AbinitBandStructureMaker.py version 1.3
=========================================
2 dataset(s) detected
Which dataset(s) do you want to use (1 to 2) ? 2
Traceback (most recent call last):
File "../band.py", line 836, in <module>
datasetkey[-1].append(float(starter[i].split()[1]))
ValueError: could not convert string to float: =
How can I fix it by step by step ?
Mutta
Physics Thailand