Note
Go to the end to download the full example code.
Convergence studies of e-DOS wrt ngkpt
This examples shows how to build a Flow to compute the band structure and the electron DOS of MgB2 with different k-point samplings.
import os
import sys
import abipy.data as abidata
import abipy.abilab as abilab
from abipy import flowtk
def make_scf_nscf_inputs(structure, pseudos, paral_kgb=1):
"""return GS, NSCF (band structure), and DOSes input."""
multi = abilab.MultiDataset(structure, pseudos=pseudos, ndtset=5)
# Global variables
multi.set_vars(
ecut=10,
nband=11,
timopt=-1,
occopt=4, # Marzari smearing
tsmear=0.03,
paral_kgb=paral_kgb,
)
# Dataset 1 (GS run)
multi[0].set_kmesh(ngkpt=[8, 8, 8], shiftk=structure.calc_shiftk())
multi[0].set_vars(tolvrs=1e-6)
# Dataset 2 (NSCF Band Structure)
multi[1].set_kpath(ndivsm=6)
multi[1].set_vars(tolwfr=1e-12)
# Dos calculations with increasing k-point sampling.
for i, nksmall in enumerate([4, 8, 16]):
multi[i+2].set_vars(
iscf=-3, # NSCF calculation
ngkpt=structure.calc_ngkpt(nksmall),
shiftk=[0.0, 0.0, 0.0],
tolwfr=1.0e-10,
)
# return GS, NSCF (band structure), DOSes input.
return multi.split_datasets()
def build_flow(options):
# Working directory (default is the name of the script with '.py' removed and "run_" replaced by "flow_")
if not options.workdir:
options.workdir = os.path.basename(sys.argv[0]).replace(".py", "").replace("run_", "flow_")
structure = abidata.structure_from_ucell("MgB2")
# Get pseudos from a table.
table = abilab.PseudoTable(abidata.pseudos("12mg.pspnc", "5b.pspnc"))
pseudos = table.get_pseudos_for_structure(structure)
inputs = make_scf_nscf_inputs(structure, pseudos)
scf_input, nscf_input, dos_inputs = inputs[0], inputs[1], inputs[2:]
return flowtk.bandstructure_flow(options.workdir, scf_input, nscf_input,
dos_inputs=dos_inputs, manager=options.manager)
# This block generates the thumbnails in the AbiPy gallery.
# You can safely REMOVE this part if you are using this script for production runs.
if os.getenv("READTHEDOCS", False):
__name__ = None
import tempfile
options = flowtk.build_flow_main_parser().parse_args(["-w", tempfile.mkdtemp()])
build_flow(options).graphviz_imshow()
@flowtk.flow_main
def main(options):
"""
This is our main function that will be invoked by the script.
flow_main is a decorator implementing the command line interface.
Command line args are stored in `options`.
"""
return build_flow(options)
if __name__ == "__main__":
sys.exit(main())
Run the script with:
run_mgb2_edoses.py -s
then use:
abirun.py flow_mgb2_edoes ebands
to get info about the electronic properties:
KS electronic bands:
nsppol nspinor nspden nkpt nband nelect fermie formula natom \
w0_t0 1 1 1 40 11 8.0 7.615 Mg1 B2 3
w0_t1 1 1 1 97 11 8.0 7.615 Mg1 B2 3
w0_t2 1 1 1 15 11 8.0 7.701 Mg1 B2 3
w0_t3 1 1 1 80 11 8.0 7.629 Mg1 B2 3
w0_t4 1 1 1 432 11 8.0 7.626 Mg1 B2 3
angle0 angle1 angle2 a b c volume abispg_num \
w0_t0 90.0 90.0 120.0 3.086 3.086 3.523 29.056 191
w0_t1 90.0 90.0 120.0 3.086 3.086 3.523 29.056 191
w0_t2 90.0 90.0 120.0 3.086 3.086 3.523 29.056 191
w0_t3 90.0 90.0 120.0 3.086 3.086 3.523 29.056 191
w0_t4 90.0 90.0 120.0 3.086 3.086 3.523 29.056 191
scheme occopt tsmear_ev \
w0_t0 cold smearing of N. Marzari with minimization ... 4 0.816
w0_t1 cold smearing of N. Marzari with minimization ... 4 0.816
w0_t2 cold smearing of N. Marzari with minimization ... 4 0.816
w0_t3 cold smearing of N. Marzari with minimization ... 4 0.816
w0_t4 cold smearing of N. Marzari with minimization ... 4 0.816
bandwidth_spin0 fundgap_spin0 dirgap_spin0 task_class \
w0_t0 12.452 0.031 0.609 ScfTask
w0_t1 12.441 0.077 0.399 NscfTask
w0_t2 12.368 0.415 1.680 NscfTask
w0_t3 12.510 0.069 0.390 NscfTask
w0_t4 12.506 0.033 0.283 NscfTask
ncfile node_id status
w0_t0 flow_mgb2_edoses/w0/t0/outdata/out_GSR.nc 241032 Completed
w0_t1 flow_mgb2_edoses/w0/t1/outdata/out_GSR.nc 241033 Completed
w0_t2 flow_mgb2_edoses/w0/t2/outdata/out_GSR.nc 241034 Completed
w0_t3 flow_mgb2_edoses/w0/t3/outdata/out_GSR.nc 241035 Completed
w0_t4 flow_mgb2_edoses/w0/t4/outdata/out_GSR.nc 241036 Completed
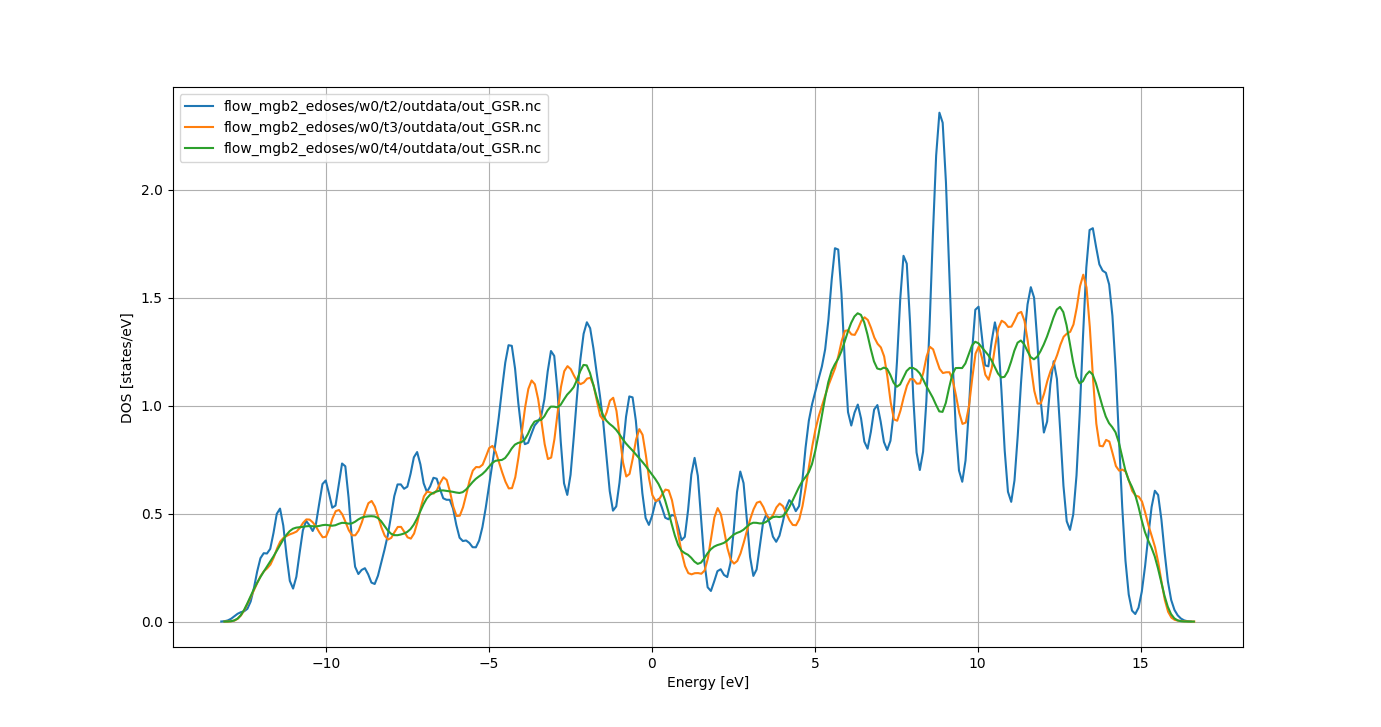
Our main goal is to analyze the convergence of the DOS wrt to the k-point sampling.
As we know that w0_t2, w0_t3` and w0_t4 are DOS calculations, we can
build a GSR robot for these tasks with:
abirun.py flow_mgb2_edoses/ ebands -nids=241034,241035,241036
then, inside the ipython shell, one can use:
In [1]: %matplotlib
In [2]: robot.combiplot_edos()
to plot the electronic DOS obtained with different number of k-points in the IBZ.
Total running time of the script: (0 minutes 0.337 seconds)