Tasks, Workflows and Flow#
In this notebook we discuss some of the basic concepts used in AbiPy to automate ab-initio calculations. In particular we will focus on the following three objects:
TaskWorkFlow
The Task represent the most elementary step of the automatic workflow.
Roughly speaking, it corresponds to the execution of a single Abinit calculation without multiple datasets.
From the point of view of AbiPy, a calculation consists of a set of Tasks that are connected by dependencies.
Each task has a list of files that are needed to start the calculation,
and a list of files that are produced at the end of the run.
Some of the input files needed by a Task must be provided by the user in the form of Abinit input variables
(e.g. crystalline structure, pseudopotentials), other inputs may be produced by other tasks.
When a Task B requires the output file DEN of another task A,
we say that B depends on A through a DEN file, and we express this dependency with the dictionary:
B_deps = {A: "DEN"}
To clarify this point, let’s take a standard KS band structure calculation as an example.
In this case, we have an initial ScfTask that solves the KS equations self-consistently to produce a DEN file.
The density is then used by a second NscfTask to compute a band structure on an arbitrary list of \(k\)-points.
The NscfTask has therefore a dependency on the ScfTask in the sense that it cannot be executed
until the ScfTask is completed and the DEN file is produced by the ScfTask.
Now that we have clarified the concept of Task, we can finally turn to Works and Flow.
The Work can be seen as a list of Tasks, while the Flow is essentially a list of Work objects.
Works are usually used to group tasks that are connected to each other.
Flows are the final objects that are executed.
The Flow provides an easy-to-use high-level API to perform common operations like launching
the actual jobs, checking the status of the Tasks, correcting problems etc.
AbiPy provides several tools to generate Flows for typical calculations, so called factory functions.
This means that you do not need to understand all the technical details of the python implementation.
In many cases, indeed, we already provide some kind of Work or Flow that automates
the whole calculation, and you only need to provide the correct list of input files.
This list, obviously, must be consistent with the kind Flow/Work you are using.
For instance, you should not pass a list of inputs for performing a band structure calculation to a Work
that is expected to compute phonons with DFPT!
All the Works and the Tasks of a flow are created and executed inside the working directory (workdir).
This is usually specified by the user during the creation of the Flow object.
AbiPy creates the workdir of the different Works/Tasks when the Flow is executed for the first time.
Each Task contains a set of input variables that will be used to generate the Abinit input file.
This input must be provided by the user during the creation of the Task.
Fortunately, AbiPy provides an object named AbinitInput to facilitate the creation of such input.
Once you have an AbinitInput, you can create the corresponding Task with the (pseudo) code:
new_task = Task(abinit_input_object)
The Task provides several methods for monitoring the status of the calculation and post-processing the results.
Note that the concept of dependency is not limited to files.
All the Tasks in the flow are connected and can interact with each other.
This allows programmers to implements python functions that will be invoked by the framework at run time.
For example, one can implement a Task that fetches the relaxed structure
from a previous Task and use this configuration to start a DFPT calculation.
In the next paragraph, we discuss how to construct a Flow for band-structure calculations
with a high-level interface that only requires the specifications of the input files.
This example allows us to discuss the most important methods of the Flow.
Building a Flow for band structure calculations#
Let’s start by creating a function that produces two input files. The first input is a standard self-consistent ground-state run. The second input uses the density produced in the first run to perform a non self-consistent band structure calculation.
# This line configures matplotlib to show figures embedded in the notebook.
# Replace `inline` with `notebook` in classic notebook
%matplotlib inline
# Option available in jupyterlab. See https://github.com/matplotlib/jupyter-matplotlib
#%matplotlib widget
import warnings
warnings.filterwarnings("ignore") # to get rid of deprecation warnings
from abipy import abilab
import os
import abipy.flowtk as flowtk
import abipy.data as abidata
def make_scf_nscf_inputs():
"""Build ands return the input files for the GS-SCF and the GS-NSCF tasks."""
multi = abilab.MultiDataset(structure=abidata.cif_file("si.cif"),
pseudos=abidata.pseudos("14si.pspnc"), ndtset=2)
# Set global variables (dataset1 and dataset2)
multi.set_vars(ecut=6, nband=8)
# Dataset 1 (GS-SCF run)
multi[0].set_kmesh(ngkpt=[8,8,8], shiftk=[0,0,0])
multi[0].set_vars(tolvrs=1e-6)
# Dataset 2 (GS-NSCF run on a k-path)
kptbounds = [
[0.5, 0.0, 0.0], # L point
[0.0, 0.0, 0.0], # Gamma point
[0.0, 0.5, 0.5], # X point
]
multi[1].set_kpath(ndivsm=6, kptbounds=kptbounds)
multi[1].set_vars(tolwfr=1e-12)
# Return two input files for the GS and the NSCF run
scf_input, nscf_input = multi.split_datasets()
return scf_input, nscf_input
Once we have our two input files, we pass them to the factory function bandstructure_flow that returns our Flow.
scf_input, nscf_input = make_scf_nscf_inputs()
workdir = "/tmp/hello_bands"
flow = flowtk.bandstructure_flow(workdir, scf_input, nscf_input)
bandstructure_flow took care of creating the correct dependency between the two tasks.
The NscfTask, indeed, depends on the ScfTask in w0/t0, whereas the ScfTask has no dependency:
flow.get_graphviz()
Note
Note that we have not used getden2 = -1 in the second dataset since AbiPy knows how to connect the two Tasks.
So no need for get* or ird* variables with Abipy.
Just specify the correct dependency and python will do the rest!
To have useful information on the status of the flow, one uses:
flow.show_status()
Work #0: <BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0>, Finalized=False
+--------+-------------+---------+--------------+------------+----------+-----------------+--------+-----------+
| Task | Status | Queue | MPI|Omp|Gb | Warn|Com | Class | Sub|Rest|Corr | Time | Node_ID |
+========+=============+=========+==============+============+==========+=================+========+===========+
| w0_t0 | Initialized | None | 1| 1|2.0 | NA|NA | ScfTask | (0, 0, 0) | None | 275 |
+--------+-------------+---------+--------------+------------+----------+-----------------+--------+-----------+
| w0_t1 | Initialized | None | 1| 1|2.0 | NA|NA | NscfTask | (0, 0, 0) | None | 276 |
+--------+-------------+---------+--------------+------------+----------+-----------------+--------+-----------+
Meaning of the different columns:
Task: short name of the task (usually w[index_of_work_in_flow]_t[index_of_task_in_work]
Status: Status of the task
Queue: QueueName@Job identifier returned by the resource manager when the task is submitted
(MPI|Omp|Gb): Number of MPI procs, OMP threads, and memory per MPI proc
(Warn|Com): Number of Error/Warning/Comment messages found in the ABINIT log
Class: The class of the
Task(Sub|Rest|Corr): Number of (submissions/restart/AbiPy corrections) performed
Node_ID : identifier of the task, used to select tasks or works in python code or
abirun.py
Both Flow and Work are iterable.
Iterating on a Flow gives Work objects, whereas
iterating over a Work gives the Tasks inside that particular Work.
for work in flow:
for task in work:
print(task)
Flows and Works are containers, and we can select items in these containers
with the syntax: flow[start:stop] or work[start:stop].
This means that the previous loop is equivalent to the much more verbose version:
for i in range(len(flow)):
work = flow[i]
for t in range(len(work):
print(work[t])
At this point it should not be so difficult to understand that:
flow[0][0]
gives the first task in the first work of the flow while
flow[-1][-1]
selects the last Task in the last Work.
In several cases, we only need to iterate over a flat list of tasks without caring about the works.
In this case, we can use:
for task in flow.iflat_tasks():
print(task)
to iterate over all Tasks in the Flow.
How to build and run the Flow#
The flow is still in memory and no file has been produced. In order to build the workflow, use:
if os.path.isdir("/tmp/hello_bands/"):
import shutil
shutil.rmtree("/tmp/hello_bands")
flow.build_and_pickle_dump()
0
This function creates the directories of the Flow:
(If you rely on MacOSX, the tree command might not be available. To fix this, see http://osxdaily.com/2016/09/09/view-folder-tree-terminal-mac-os-tree-equivalent/)
!tree /tmp/hello_bands
/tmp/hello_bands
├── __AbinitFlow__.pickle
├── _fix_flow.py
├── indata
├── outdata
├── tmpdata
└── w0
├── indata
├── outdata
├── t0
│ ├── abipy_meta.json
│ ├── indata
│ ├── job.sh
│ ├── outdata
│ ├── run.abi
│ └── tmpdata
├── t1
│ ├── abipy_meta.json
│ ├── indata
│ ├── job.sh
│ ├── outdata
│ ├── run.abi
│ └── tmpdata
└── tmpdata
16 directories, 8 files
Let’s have a look at the files/directories associated to the first work (flow[0]):
flow[0].get_graphviz_dirtree()
w0 is the directory containing the input files of the first workflow (well, we have only one workflow in our example).
t0 and t1 contain the input files needed to run the SCF and the NSC run, respectively.
You might have noticed that each Task directory present the same structure:
run.abi: Input file
run.files: Files file
job.sh: Submission script
outdata: Directory containing output data files
indata: Directory containing input data files
tmpdata: Directory with temporary files
Danger
AbinitFlow.pickle is the pickle file used to save the status of Flow. Don’t touch it!
An Abinit Task has an AbinitInput which in turn has a Structure:
flow[0][0].input
#### SECTION: basic
##############################################
ecut 6
nband 8
ngkpt 8 8 8
kptopt 1
nshiftk 1
shiftk 0 0 0
tolvrs 1e-06
##############################################
#### SECTION: files
##############################################
indata_prefix indata/in
tmpdata_prefix tmpdata/tmp
outdata_prefix outdata/out
pseudos /home/runner/work/abipy_book/abipy_book/abipy/abipy/data/pseudos/14si.pspnc
##############################################
#### STRUCTURE
##############################################
natom 2
ntypat 1
typat 1 1
znucl 14
xred
0.0000000000 0.0000000000 0.0000000000
0.2500000000 0.2500000000 0.2500000000
acell 1.0 1.0 1.0
rprim
6.3285005287 0.0000000000 3.6537614838
2.1095001762 5.9665675181 3.6537614838
0.0000000000 0.0000000000 7.3075229676
flow[0][0].input.structure
Structure Summary
Lattice
abc : 3.86697462 3.86697462 3.86697462
angles : 59.99999999999999 59.99999999999999 59.999999999999986
volume : 40.88829179346891
A : np.float64(3.3488982567096763) np.float64(0.0) np.float64(1.9334873100000005)
B : np.float64(1.1162994189032256) np.float64(3.1573715557642927) np.float64(1.9334873100000005)
C : np.float64(0.0) np.float64(0.0) np.float64(3.86697462)
pbc : True True True
PeriodicSite: Si1 (Si) (0.0, 0.0, 0.0) [0.0, 0.0, 0.0]
PeriodicSite: Si2 (Si) (1.116, 0.7893, 1.933) [0.25, 0.25, 0.25]
for p in flow[0][0].input.pseudos:
print(p)
<NcAbinitPseudo: 14si.pspnc>
summary: Troullier-Martins psp for element Si Thu Oct 27 17:31:21 EDT 1994
number of valence electrons: 4.0
maximum angular momentum: d
angular momentum for local part: d
XC correlation: LDA_XC_TETER93
supports spin-orbit: False
radius for non-linear core correction: 1.80626423934776
hint for low accuracy: ecut: 0.0, pawecutdg: 0.0
hint for normal accuracy: ecut: 0.0, pawecutdg: 0.0
hint for high accuracy: ecut: 0.0, pawecutdg: 0.0
Let’s print the value of kptopt for all tasks in our flow with:
print([task.input["kptopt"] for task in flow.iflat_tasks()])
[1, -2]
that, in this particular case, gives the same result as:
print([task.input["kptopt"] for task in flow[0]])
[1, -2]
Executing a Flow#
The Flow can be executed with two different approaches: a programmatic interface based
on flow.make_scheduler or the abirun.py script.
In this section, we discuss the first approach because it plays well with the jupyter notebook.
Note however that abirun.py is highly recommended especially when running non-trivial calculations.
flow.make_scheduler().start()
Using scheduler v >= 3.0.0
[Thu Jan 15 12:19:09 2026] Number of launches: 1
Work #0: <BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0>, Finalized=False
+--------+-------------+------------+--------------+------------+----------+-----------------+----------+-----------+
| Task | Status | Queue | MPI|Omp|Gb | Warn|Com | Class | Sub|Rest|Corr | Time | Node_ID |
+========+=============+============+==============+============+==========+=================+==========+===========+
| w0_t0 | Submitted | 3971@githu | 2| 1|2.0 | NA|NA | ScfTask | (1, 0, 0) | 0:00:00Q | 275 |
+--------+-------------+------------+--------------+------------+----------+-----------------+----------+-----------+
| w0_t1 | Initialized | None | 1| 1|2.0 | NA|NA | NscfTask | (0, 0, 0) | None | 276 |
+--------+-------------+------------+--------------+------------+----------+-----------------+----------+-----------+
nbdbuf is not specified in input, using `nbdbuf = 2 * nspinor` that may be too small!"
As a consequence, the NSCF cycle may have problems to converge the last bands within nstep iterations.
To avoid this problem specify nbdbuf in the input file and adjust nband accordingly.
[Thu Jan 15 12:19:14 2026] Number of launches: 1
Work #0: <BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0>, Finalized=False
+--------+-----------+------------+--------------+------------+----------+-----------------+----------+-----------+
| Task | Status | Queue | MPI|Omp|Gb | Warn|Com | Class | Sub|Rest|Corr | Time | Node_ID |
+========+===========+============+==============+============+==========+=================+==========+===========+
| w0_t0 | Completed | 3971@githu | 2| 1|2.0 | 2| 2 | ScfTask | (1, 0, 0) | 0:00:01R | 275 |
+--------+-----------+------------+--------------+------------+----------+-----------------+----------+-----------+
| w0_t1 | Submitted | 3991@githu | 2| 1|2.0 | NA|NA | NscfTask | (1, 0, 0) | 0:00:00Q | 276 |
+--------+-----------+------------+--------------+------------+----------+-----------------+----------+-----------+
Work #0: <BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0>, Finalized=True
Finalized works are not shown. Use verbose > 0 to force output.
all_ok reached
Calling flow.finalize() ...
Submitted on: Thu Jan 15 12:19:09 2026
Completed on: Thu Jan 15 12:19:19 2026
Elapsed time: 0:00:10.007245
Flow completed successfully
0
The flow keeps track of the different actions performed by the python code:
flow.show_history()
================================================================================
<BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0>
================================================================================
[Thu Jan 15 12:19:14 2026] In on_ok with sender <ScfTask, node_id=275, workdir=../../../../../../tmp/hello_bands/w0/t0>
[Thu Jan 15 12:19:19 2026] In on_ok with sender <NscfTask, node_id=276, workdir=../../../../../../tmp/hello_bands/w0/t1>
[Thu Jan 15 12:19:19 2026] Finalized set to True
[Thu Jan 15 12:19:19 2026] Work <BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0> is finalized and broadcasts signal S_OK
[Thu Jan 15 12:19:19 2026] Node <BandStructureWork, node_id=274, workdir=../../../../../../tmp/hello_bands/w0> broadcasts signal Completed
===============================================================================
=== <ScfTask, node_id=275, workdir=../../../../../../tmp/hello_bands/w0/t0> ===
===============================================================================
[Thu Jan 15 12:19:09 2026] Status changed to Ready. msg: Status set to Ready
[Thu Jan 15 12:19:09 2026] Setting input variables: {'autoparal': 1, 'max_ncpus': 2, 'mem_test': 0, 'chkparal': 0}
[Thu Jan 15 12:19:09 2026] Old values: {'autoparal': None, 'max_ncpus': None, 'mem_test': None, 'chkparal': None}
[Thu Jan 15 12:19:09 2026] Setting input variables: {'npimage': 1, 'npkpt': 2, 'npspinor': 1, 'npfft': 1, 'npband': 1, 'bandpp': 1}
[Thu Jan 15 12:19:09 2026] Old values: {'npimage': None, 'npkpt': None, 'npspinor': None, 'npfft': None, 'npband': None, 'bandpp': None}
[Thu Jan 15 12:19:09 2026] Status changed to Initialized. msg: finished autoparal run
[Thu Jan 15 12:19:09 2026] Submitted with MPI=2, Omp=1, Memproc=2.0 [GB] Submitted to queue
[Thu Jan 15 12:19:14 2026] Task completed status set to OK based on abiout
[Thu Jan 15 12:19:14 2026] Finalized set to True
[Thu Jan 15 12:19:14 2026] Node <ScfTask, node_id=275, workdir=../../../../../../tmp/hello_bands/w0/t0> broadcasts signal Completed
================================================================================
=== <NscfTask, node_id=276, workdir=../../../../../../tmp/hello_bands/w0/t1> ===
================================================================================
[Thu Jan 15 12:19:14 2026] Status changed to Ready. msg: Status set to Ready
[Thu Jan 15 12:19:14 2026] Need path `/tmp/hello_bands/w0/t0/outdata/out_DEN` with extension: `DEN`
[Thu Jan 15 12:19:14 2026] Linking path /tmp/hello_bands/w0/t0/outdata/out_DEN --> /tmp/hello_bands/w0/t1/indata/in_DEN
[Thu Jan 15 12:19:14 2026]
nbdbuf is not specified in input, using `nbdbuf = 2 * nspinor` that may be too small!"
As a consequence, the NSCF cycle may have problems to converge the last bands within nstep iterations.
To avoid this problem specify nbdbuf in the input file and adjust nband accordingly.
[Thu Jan 15 12:19:14 2026] Setting input variables: {'ngfft': array([18, 18, 18])}
[Thu Jan 15 12:19:14 2026] Old values: {'ngfft': None}
[Thu Jan 15 12:19:14 2026] Adding connecting vars {'irdden': 1}
[Thu Jan 15 12:19:14 2026] Setting input variables: {'irdden': 1}
[Thu Jan 15 12:19:14 2026] Old values: {'irdden': None}
[Thu Jan 15 12:19:14 2026] Setting input variables: {'autoparal': 1, 'max_ncpus': 2, 'mem_test': 0, 'chkparal': 0}
[Thu Jan 15 12:19:14 2026] Old values: {'autoparal': None, 'max_ncpus': None, 'mem_test': None, 'chkparal': None}
[Thu Jan 15 12:19:14 2026] Setting input variables: {'npimage': 1, 'npkpt': 2, 'npspinor': 1, 'npfft': 1, 'npband': 1, 'bandpp': 1}
[Thu Jan 15 12:19:14 2026] Old values: {'npimage': None, 'npkpt': None, 'npspinor': None, 'npfft': None, 'npband': None, 'bandpp': None}
[Thu Jan 15 12:19:14 2026] Status changed to Initialized. msg: finished autoparal run
[Thu Jan 15 12:19:14 2026] Submitted with MPI=2, Omp=1, Memproc=2.0 [GB] Submitted to queue
[Thu Jan 15 12:19:19 2026] Task completed status set to OK based on abiout
[Thu Jan 15 12:19:19 2026] Finalized set to True
[Thu Jan 15 12:19:19 2026] Node <NscfTask, node_id=276, workdir=../../../../../../tmp/hello_bands/w0/t1> broadcasts signal Completed
================================================================================
======== <Flow, node_id=273, workdir=../../../../../../tmp/hello_bands> ========
================================================================================
[Thu Jan 15 12:19:19 2026] Calling flow.finalize.
[Thu Jan 15 12:19:19 2026] Finalized set to True
stty: 'standard input': Inappropriate ioctl for device
If you read the logs carefully, you will realize that in the first iteration of the scheduler,
only the ScfTask is executed because the second task depends on it.
After the initial submission, the scheduler starts to monitor all the tasks in the flow.
When the ScfTask completes, the dependency of the NscfTask is fulfilled and a new submission takes place.
Once the second task completes, the scheduler calls flow.finalize
to execute (optional) logic that is supposed to be executed to perform some sort of cleanup or post-processing.
At this point, all the tasks in the flow are completed and the scheduler exits.
Now we can have a look at the different output files produced by the flow with:
flow[0].get_graphviz_dirtree()
or list only the files with a given extension:
flow.listext("GSR.nc")
Found 2 files with extension `GSR.nc` produced by the flow
File Size [Mb] Node_ID Node Class
---------------------------------------------------------- ----------- --------- ------------
../../../../../../tmp/hello_bands/w0/t0/outdata/out_GSR.nc 9.63 275 ScfTask
../../../../../../tmp/hello_bands/w0/t1/outdata/out_GSR.nc 9.62 276 NscfTask
The nice thing about the flow is that the object knows how to locate and interpret the different input/output files produced by Abinit. As a consequence, it is very easy to expose the AbiPy post-processing tools with a easy-to-use API in which only tasks/works/flow plus a very few input arguments are required.
Let’s call, for instance, the inspect method to plot the self-consistent cycles:
flow.inspect(tight_layout=True);
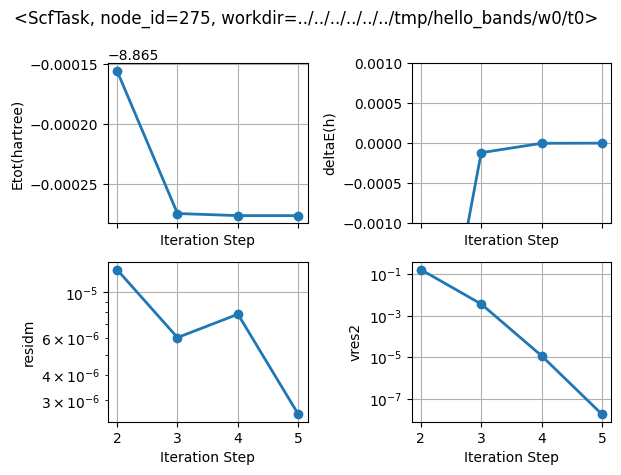
Task <NscfTask, node_id=276, workdir=../../../../../../tmp/hello_bands/w0/t1> does not provide an inspect method
In the other AbiPy tutorials, we have explained how to use abiopen to create python objects from netcdf files. Well, the same code can be reused with the flow. It is just a matter of replacing
with abiopen(filepath) as gsr:
with
with task.open_gsr() as gsr:
Note that there is no need to specify the file path when you use the task-based API, because
the Task knows how to locate its GSR.nc output.
Let’s do some practice…
with flow[0][0].open_gsr() as gsr:
ebands_kmesh = gsr.ebands
with flow[0][1].open_gsr() as gsr:
gsr.ebands.plotly_with_edos(ebands_kmesh.get_edos(), with_gaps=True);
More on Works, Tasks and dependencies#
In the previous example, we have constructed a workflow for band structure calculations starting from two input files and the magic line
flow = flowtk.bandstructure_flow(workdir, scf_input, nscf_input)
Now it is the right time to explain in more details the syntax and the API used in AbiPy
to build a flow with dependencies.
Let’s try to build a Flow from scratch and use graphviz after each step to show what’s happening.
We start with an empty flow in the hello_flow directory:
hello_flow = flowtk.Flow(workdir="hello_flow")
hello_flow.get_graphviz()
Now we add a new Task by just passing an AbinitInput for SCF calculations:
hello_flow.register_scf_task(scf_input, append=True)
hello_flow.get_graphviz()
Now the tricky part.
We want to register a NSCF calculation that should depend on the scf_task in w0_t0 via the DEN file.
We can use the same API but we must specify the dependency between the two steps with the
{scf_task: "DEN"} dictionary:
hello_flow.register_nscf_task(nscf_input, deps={hello_flow[0][0]: "DEN"}, append=True)
hello_flow.get_graphviz(engine="dot")
Excellent, we managed to build our first AbiPy flow with inter-dependent tasks in just six lines
of code (including the three calls to graphviz).
Now let’s assume we want to add a second Nscf calculation (NscTask) in which we change one of the input parameters
e.g. the number of bands and that, for some reason, we really want to reuse the output WFK file
produced by w0_t1 to initialize the eigenvalue solver (obviously we still need a DEN file).
How can we express this with AbiPy?
Well, the syntax for the new deps, it’s just:
deps = {hello_flow[0][0]: "DEN", hello_flow[0][1]: "WFK"}
but we should also change the input variable nband in the nscf_input before creating
the new NscTask (remember that building a Task requires an AbinitInput object
and a list of dependencies, if any).
Now there are two ways to increase nband: the wrong way and the correct one! Let’s start from the wrong way because it’s always useful to learn from our mistakes. Let’s print some values just for the record:
t1 = flow[0][1]
print("nband in the first NscfTask:", t1.input["nband"])
nband in the first NscfTask: 8
Now let’s use the “recipe” recommended to us by the FORTRAN guru of our group:

# Just copy the previous input and change nband, it's super-easy, the FORTRAN guru said!
new_input = t1.input
new_input["nband"] = 1000
print("nband in the first NscfTask:", t1.input["nband"])
print("nband in the new input:", new_input["nband"])
nband in the first NscfTask: 1000
nband in the new input: 1000
Tada! Thanks to the trick of our beloved FORTRAN guru, we ended up with two NscfTaks with the same number of bands (1000!). Why?
Because AbinitInput is implemented internally with a dictionary, python dictionaries are mutable
and python variables are essentially references (they do not store data, actually they store the address of the data).
a = {"foo": "bar"}
b = a
c = a.copy()
a["hello"] = "world"
print("a dict:", a)
print("b dict:", b)
print("c dict:", c)
a dict: {'foo': 'bar', 'hello': 'world'}
b dict: {'foo': 'bar', 'hello': 'world'}
c dict: {'foo': 'bar'}
For a more technical explanation see here
To avoid this mistake, we need to copy the object before changing it
t1.input["nband"] = 8 # back to the old value
new_input = t1.input.new_with_vars(nband=1000) # Copy and change nband
print("nband in the first NscfTask:", t1.input["nband"])
print("nband in the new input:", new_input["nband"])
nband in the first NscfTask: 8
nband in the new input: 1000
Now we can finally add the second NscfTask with 1000 bands:
hello_flow.register_nscf_task(new_input, deps={hello_flow[0][0]: "DEN", hello_flow[0][1]: "WFK"}, append=True)
hello_flow.get_graphviz(engine="dot")
print([task.input["nband"] for task in hello_flow.iflat_tasks()])
[8, 8, 1000]
Note that AbiPy dependencies can also be fulfilled with external files that are already available
when the flow is constructed.
There is no change in the syntax we’ve used so far.
It is just a matter of using the absolute path to the DEN file as keyword of the dictionary instead of a Task.
Let’s start with a new Flow to avoid confusion and create a NscfTask that will start from a pre-computed DEN file.
flow_with_file = flowtk.Flow(workdir="flow_with_file")
den_filepath = abidata.ref_file("si_DEN.nc")
flow_with_file.register_nscf_task(nscf_input, deps={den_filepath: "DEN"})
flow_with_file.get_graphviz(engine="dot")
A call to new_with_vars inside a python for loop is all we need to add other two NscfTasks
with different nband, all starting from the same DEN file:
for nband in [10, 20]:
flow_with_file.register_nscf_task(nscf_input.new_with_vars(nband=nband),
deps={den_filepath: "DEN"}, append=False)
print([task.input["nband"] for task in flow_with_file.iflat_tasks()])
flow_with_file.get_graphviz()
[8, 10, 20]
At this point, you may ask why we need Works since all the examples presented so far mainly involve the Flow object.
The answer is that Works allow us to encapsulate reusable logic in magic boxes that can perform lot of useful work.
These boxes can then be connected together to generate more complicated workflows.
We have already encountered the BandStructureWork at the beginning of this lesson
and now it is time to introduce another fancy animal of the AbiPy zoo, the PhononWork.
abilab.print_doc(flowtk.PhononWork)
class PhononWork(Work, MergeDdb):
"""
This work consists of nirred Phonon tasks where nirred is
the number of irreducible atomic perturbations for a given set of q-points.
It provides the callback method (on_all_ok) that calls mrgddb and mrgdv to merge
all the partial DDB (POT) files produced.
The two files are available in the output directory of the Work.
.. rubric:: Inheritance Diagram
.. inheritance-diagram:: PhononWork
"""
abilab.print_doc(flowtk.PhononWork.from_scf_task)
@classmethod
def from_scf_task(cls, scf_task: ScfTask,
qpoints, is_ngqpt=False, with_becs=False,
with_quad=False, with_flexoe=False, with_dvdb=True,
tolerance=None, ddk_tolerance=None, ndivsm=0, qptopt=1,
prtwf=-1, prepgkk=0, manager=None) -> PhononWork:
"""
Construct a `PhononWork` from a |ScfTask| object.
The input file for phonons is automatically generated from the input of the ScfTask.
Each phonon task depends on the WFK file produced by the `scf_task`.
Args:
scf_task: |ScfTask| object.
qpoints: q-points in reduced coordinates. Accepts single q-point, list of q-points
or three integers defining the q-mesh if `is_ngqpt`.
is_ngqpt: True if `qpoints` should be interpreted as divisions instead of q-points.
with_becs: Activate calculation of Electric field and Born effective charges.
with_quad: Activate calculation of dynamical quadrupoles. Require `with_becs`
Note that only selected features are compatible with dynamical quadrupoles.
Please consult <https://docs.abinit.org/topics/longwave/>
with_flexoe: True to activate computation of flexoelectric tensor. Require `with_becs`
with_dvdb: True to merge POT1 files associated to atomic perturbations in the DVDB file
at the end of the calculation
tolerance: dict {"varname": value} with the tolerance to be used in the phonon run.
None to use AbiPy default.
ddk_tolerance: dict {"varname": value} with the tolerance used in the DDK run if with_becs.
None to use AbiPy default.
ndivsm: If different from zero, activate computation of electron band structure.
if > 0, it's the number of divisions for the smallest segment of the path (Abinit variable).
if < 0, it's interpreted as the pymatgen `line_density` parameter in which the number of points
in the segment is proportional to its length. Typical value: -20.
This option is the recommended one if the k-path contains two high symmetry k-points that are very close
as ndivsm > 0 may produce a very large number of wavevectors.
if 0, deactivate band structure calculation.
qptopt: Option for the generation of q-points. Default: 1 i.e. use msym spatial symmetries + TR.
prtwf: Controls the output of the first-order WFK.
By default we set it to -1 when q != 0 so that AbiPy is still able
to restart the DFPT task if the calculation is not converged (worst case scenario)
but we avoid the output of the 1-st order WFK if the calculation converged successfully.
Non-linear DFPT tasks should not be affected since they assume q == 0.
prepgkk: option to compute only the irreducible preturbations or all perturbations
for the chosen set of q-point. Default: 0 (irred. pret. only)
manager: |TaskManager| object.
"""
The docstring seems to suggest that if I have a scf_task, I can construct a magic box
to compute phonons but wait, I already have such a task!
Actually I already have another magic box to compute the electronic band structure
and it would be really great if I could compute the electronic and vibrational properties in a single flow.
Let’s connect the two boxes together with:
# Create new flow.
ph_flow = flowtk.Flow(workdir="phflow")
# Band structure (SCF + NSCF)
bands_work = flowtk.BandStructureWork(scf_input, nscf_input, dos_inputs=None)
ph_flow.register_work(bands_work)
# Build second work from scf_task.
scf_task = bands_work[0]
ph_work = flowtk.PhononWork.from_scf_task(scf_task, [2, 2, 2], is_ngqpt=True, tolerance=None)
ph_flow.register_work(ph_work)
ph_flow.get_graphviz()
Now it turns out that the PhononWork merges all the DDB files produced by its PhononTask
and put this final output file in its outdir.
So from the AbiPy perspective, a PhononWork is not that different from a ScfTask that produces e.g. a DEN file.
This means that we can connect other magic boxes to our PhononWork e.g. a set of EPhTasks that
require a DDB file and another input file with the DFPT potentials
(DVDB, merged by PhononWork similarly to what is done for the DDB).
# EPH tasks require 3 input files (WFK, DDB, DVDB)
eph_deps = {ph_flow[0][1]: "WFK", ph_work: ["DDB", "DVDB"]}
for i, ecut in enumerate([2, 3, 4]):
ph_flow.register_eph_task(nscf_input.new_with_vars(ecut=ecut), deps=eph_deps, append=(i != 0))
ph_flow.get_graphviz()
This explains why in AbiPy we have this classification in terms of Tasks/Works/Flows.
As a consequence, we can implement highly specialized Works/Tasks to
solve specific problems and then connect all these nodes together.
Just 11 lines of code to get electrons + phonons + (electrons + phononons)!
But wait, did you see the gorilla?
from IPython.display import YouTubeVideo
YouTubeVideo("vJG698U2Mvo")
There’s indeed a bug in the last step. The connections among the nodes are OK but we made
a mistake while creating the EPhTasks with:
ph_flow.register_eph_task(nscf_input.new_with_vars(ecut=ecut), ...)
because we passed an input for a standard band structure calculation to something that is supposed to deal with E-PH interaction.
This essentially to stress that the AbiPy Flow, by design, does not try to validate your input to make sure
it is consistent with the workflow logic.
This is done on purpose for two reasons:
Expert users should be able to customize/tune their input files and validating all the possible cases in python is not trivial
Only Abinit (and God) knows at run-time if your input file makes sense and we can’t reimplement the same logic in python
At this point, you may wonder why we have so many different Abipy Tasks (ScfTask, NscfTask, RelaxTask, PhononTask, EPHTask …)
if there’s no input validation when we create them…
The answer is that we need all these subclasses to implement extra logic that is specific to that particular calculation. Abipy, indeed, is not just submitting jobs. It also monitors the evolution of the calculation and execute pre-defined code to fix run-time problems (and these problems are calculation specific). An example will help clarify this point.
Restarting jobs is one of the typical problem encountered in ab-initio calculations
and restarting a RelaxTask requires a different logic from e.g. restarting a ScfTask.
In the case of a ScfTask we only need to use the output WFK (DEN) of the previous execution
as input of the restarted job while a RelaxTask must also reuse the (unconverged) final structure
of the previous job to be effective and avoid a possibly infinite loop.
In a nutshell, when you are using a particular Task/Work class you are telling AbiPy how to handle possible
problems at run-time and you are also specifying the actions that should be performed
at the beginning/end of the execution.
Abirun.py#
Executing
flow.make_scheduler().start()
inside a jupyter notebook is handy if you are dealing with small calculations that require few seconds or minutes. This approach, however, is unpractical when you have large flows or big calculations requiring hours or days, even on massively parallel machines. In this case, indeed, one would like to run the scheduler in a separate process in the background so that the scheduler is not killed when the jupyter server is closed.
To start the scheduler in a separate process, use the abirun.py script.
The syntax is:
abirun.py flow_workdir COMMAND
where flow_workdir is the directory containing the Flow
(the directory with the pickle file) and command selects the operation to be performed.
Typical examples:
abirun.py /tmp/hello_bands status
checks the status of the Flow and print the results to screen while
nohup abirun.py /tmp/hello_bands scheduler > sched.log 2> sched.err &
starts the scheduler in the background redirecting the standard output to file sched.log
Important
nohup is a standard Unix tool. The command make the scheduler immune
to hangups so that you can close the shell session without killing the scheduler.
This brings us to the last and most crucial question. How do we configure AbiPy to run Abinit workflows on different architectures ranging from standard laptops to high-performance supercomputers?
Unfortunately this notebook is already quite long and these details are best covered in a technical documentation.
What should be stressed here is that the behaviour can be customized with two Yaml files.
All the information related to your environment (Abinit build, modules, resource managers, shell environment)
are read from the manager.yml configuration file, that is usually located in the directory ~/.abinit/abipy/
The options for the python scheduler responsible for job submission are given in scheduler.yml.
For a more complete description of these configuration options, please consult the TaskManager documentation. A list of configuration files for different machines and clusters is available here while the Flows HOWTO gathers answers to frequently asked questions.
Last but not least, check out our gallery of AbiPy Flows for inspiration.

