Structure object#
The AbiPy structure inherits from the pymatgen structure.
One has therefore access to all the methods and tools already available in pymatgen.
In this notebook, we mainly focus on the extensions added by AbiPy.
For the features provided by pymatgen, please consult the
official pymatgen documentation.
abipy.core.structure.Structure and pymatgen.core.structure.Structure
import warnings
warnings.filterwarnings("ignore") # to get rid of deprecation warnings
# Import abipy modules
from abipy import abilab
from abipy.abilab import Structure
import abipy.data as abidata
# Useful tools we'll need later on.
from pprint import pprint
import numpy as np
# This line configures matplotlib to show figures embedded in the notebook.
# Replace `inline` with `notebook` in classic notebook
%matplotlib inline
# Option available in jupyterlab. See https://github.com/matplotlib/jupyter-matplotlib
#%matplotlib widget
Reading a structure from file#
It is possible to initialize a structure object from different file formats:
CIF
POSCAR/CONTCAR
CHGCAR
LOCPOT,
vasprun.xml
CSSR
ABINIT Netcdf files
pymatgen’s JSON serialized structures
Note, in particular, that one can initialize the structure from the netcdf files
produced by Abinit (GSR.nc, WFK.nc, etc) as well as output files in text format
such as the Abinit input/output files or even the DDB file.
To initialize the structure from a CIF file use the from_file method:
structure = Structure.from_file(abidata.cif_file("si.cif"))
print(structure)
Full Formula (Si2)
Reduced Formula: Si
abc : 3.866975 3.866975 3.866975
angles: 60.000000 60.000000 60.000000
pbc : True True True
Sites (2)
# SP a b c
--- ---- ---- ---- ----
0 Si 0 0 0
1 Si 0.25 0.25 0.25
Note
abidata.cif_file, abidata.ref_file and abidata.pseudos
are helper function returning the absolute path of one of the reference files shipped with the AbiPy package.
In your case, you can directly pass a string or a list of strings with the path to your files.
To read the structure from an Abinit netcdf file, use:
structure = Structure.from_file(abidata.ref_file("si_nscf_GSR.nc"))
print(structure.to_string(verbose=1)) # Use to_string with verbose > 0 to get more info
Full Formula (Si2)
Reduced Formula: Si
abc : 3.866975 3.866975 3.866975
angles: 60.000000 60.000000 60.000000
Spglib space group info (magnetic symmetries not taken into account).
Spacegroup: Fd-3m (227), Hall: F 4d 2 3 -1d, Abinit spg_number: 227
Crystal_system: cubic, Lattice_type: cubic, Point_group: m-3m
Idx Symbol Reduced_Coords Wyckoff EqIdx
----- -------- -------------------------- --------- -------
0 Si +0.00000 +0.00000 +0.00000 (2a) 0
1 Si +0.25000 +0.25000 +0.25000 (2a) 0
Abinit Spacegroup: spgid: 227, num_spatial_symmetries: 48, has_timerev: True, symmorphic: True
Use to_abivars to get a python dictionary with the list of Abinit variables.
structure.to_abivars()
{'natom': 2,
'ntypat': 1,
'typat': array([1, 1]),
'znucl': [14],
'xred': array([[0. , 0. , 0. ],
[0.25, 0.25, 0.25]]),
'acell': [1.0, 1.0, 1.0],
'rprim': array([[6.32850055, 0. , 3.6537615 ],
[2.10950018, 5.96656754, 3.6537615 ],
[0. , 0. , 7.30752299]])}
and the abi_string property to get a string that can be used directly in the input file:
print(structure.abi_string)
natom 2
ntypat 1
typat 1 1
znucl 14
xred
0.0000000000 0.0000000000 0.0000000000
0.2500000000 0.2500000000 0.2500000000
acell 1.0 1.0 1.0
rprim
6.3285005521 0.0000000000 3.6537614973
2.1095001840 5.9665675402 3.6537614973
0.0000000000 0.0000000000 7.3075229946
To visualize the structure with matplotlib, use:
structure.plot();
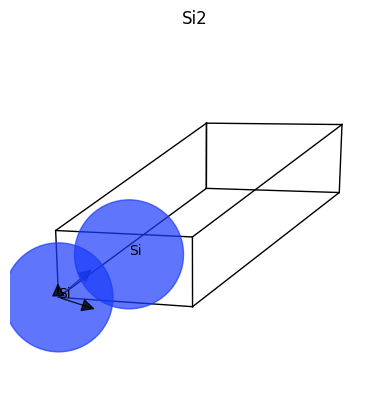
The matplotlib version is minimalistic but it plays well with jupyter notebooks. For a more advanced visualization we suggest using a specialized graphical applications. Fortunately, one can invoke external applications directly from AbiPy with e.g.
# structure.visualize("vesta")
provided VESTA is already installed on your machine and the binary can be found in $PATH.
To get a structure from the materials project database, use:
# Remember to set the env variable PMG_MAPI_KEY in your ~/.pmgrc.yaml files.
si2_mp = Structure.from_mpid("mp-149")
print(si2_mp)
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[8], line 2
1 # Remember to set the env variable PMG_MAPI_KEY in your ~/.pmgrc.yaml files.
----> 2 si2_mp = Structure.from_mpid("mp-149")
3 print(si2_mp)
File ~/work/abipy_book/abipy_book/abipy/abipy/core/structure.py:390, in Structure.from_mpid(cls, material_id)
388 # Get pytmatgen structure and convert it to abipy structure
389 from abipy.core import restapi
--> 390 with restapi.get_mprester() as rest:
391 new = rest.get_structure_by_material_id(material_id)
392 return cls.as_structure(new)
File ~/work/abipy_book/abipy_book/abipy/abipy/core/restapi.py:33, in get_mprester()
29 def get_mprester():
30 """
31 Args:
32 """
---> 33 rester = MPRester()
34 #print(f"{type(rester)=}")
35 return rester
File /usr/share/miniconda/envs/abipy/lib/python3.12/site-packages/pymatgen/ext/matproj.py:125, in MPRester.__init__(self, api_key, include_user_agent)
122 self.api_key = SETTINGS.get("PMG_MAPI_KEY", "")
124 if len(self.api_key) != 32:
--> 125 raise ValueError(
126 "Invalid or old API key. Please obtain an updated API key at https://materialsproject.org/dashboard."
127 )
129 self.preamble = SETTINGS.get("PMG_MAPI_ENDPOINT", "https://api.materialsproject.org/")
131 self.session = requests.Session()
ValueError: Invalid or old API key. Please obtain an updated API key at https://materialsproject.org/dashboard.
In some cases, we have multiple structures and we need to compare the lattice parameters.
Use dataframes_from_structures to build a pandas DataFrame:
dfs = abilab.dataframes_from_structures([structure, si2_mp], index=["CIF", "MP"])
then we can compare the lattice parameters with:
dfs.lattice
Note that all AbiPy robots have this feature built-in.
Sometimes it is much easier to build a robot directly from files
and then compare the structures with e.g. robot.get_lattice_dataframe().
Converting to other formats#
Use structure.convert(format) to get the string representation in the new format:
for fmt in ("cif", "POSCAR", "qe"):
print((" Abinit --> %s " % fmt).center(80, "*"))
print(structure.convert(fmt=fmt))
Getting info on the structure#
print(structure.reciprocal_lattice)
structure.reciprocal_lattice.matrix.T @ structure.lattice.matrix / (2 * np.pi)
# List of high-symmetry k-points.
print(structure.hsym_kpoints)
The method calc_ksampling allows one to get an efficient sampling of the Brillouin zone
by just specifying the number of divisions to be used for the smallest lattice vector of the reciprocal lattice:
pprint(structure.calc_ksampling(nksmall=10))
To get the recommended high symmetry \(k\)-path in reduced coordinates:
structure.calc_kptbounds()
The high-symmetry q-path is automatically selected assuming the structure fulfills the convention described in Setyawan2010
To visualize the Brillouin zone with matplotlib, use:
structure.plot_bz();
For the plotly version, use:
structure.plotly_bz();
Note
The name of the plotly method (if implemented) is obtained by replacing the plot verb with plotly.
To get the number of valence electrons for a given set of pseudopotentials:
structure.num_valence_electrons(pseudos=abidata.pseudos("14si.pspnc"))
To visualize the X-ray diffraction plot with pymatgen XRDCalculator, use:
structure.plot_xrd();
The abistruct.py script#
The abistruct.py script provides a handy command line interface to operate on structure objects constructed from external files. There are several options available as well an interface to the and the COD database.
To obtain the list of available commands, use:
!abistruct.py --help
Creating a GUI inside a notebook#
Several AbiPy objects provide a get_panel method that allows one to create a panel GUI
exposing some of the underlying AbiPy methods.
Similar capabilities are also available via the AbiPy GUI web app.
To build a panel GUI for a given structure use:
abilab.abipanel()
structure.get_panel()

