Note
Go to the end to download the full example code.
Lobster COHP/COP/DOS
This example shows how to analyze the output files produced by Lobster code <<http://schmeling.ac.rwth-aachen.de/cohp/>
Use abiview.py lobster DIRPATH for a command line interface.
================================= COOP File =================================
COOP: Number of energies: 401, from -14.035 to 6.015 (eV) with E_fermi set 0 (was 2.298)
has_partial_projections: True, nsppol: 1
Number of pairs: 2
[0] Ga@0 --> As@1
[1] As@1 --> Ga@0
================================= COHP File =================================
COHP: Number of energies: 401, from -14.035 to 6.015 (eV) with E_fermi set 0 (was 2.298)
has_partial_projections: True, nsppol: 1
Number of pairs: 2
[0] Ga@0 --> As@1
[1] As@1 --> Ga@0
============================== ICHOHPLIST File ==============================
Number of pairs: 2
index0 index1 type0 type1 spin average distance n_bonds pair
0 1 Ga As 0 -4.36062 2.49546 None (0, 1)
1 0 As Ga 0 -4.36062 2.49546 None (1, 0)
=============================== Lobster DOSCAR ===============================
Number of energies: 401, from -14.035 to 6.015 (eV) with E_fermi set to 0 (was 2.298)
nsppol: 1
Number of sites in projected DOS: 2
0 --> {4s, 4p_y, 4p_z, 4p_x}
1 --> {4s, 4p_y, 4p_z, 4p_x}
import os
import abipy.data as abidata
from abipy.abilab import LobsterAnalyzer
dirpath = os.path.join(abidata.dirpath, "refs", "lobster_gaas")
# Open the all the lobster files produced in directory dirpath
# with the (optional) prefix GaAs_
lobana = LobsterAnalyzer.from_dir(dirpath, prefix="GaAs_")
print(lobana)
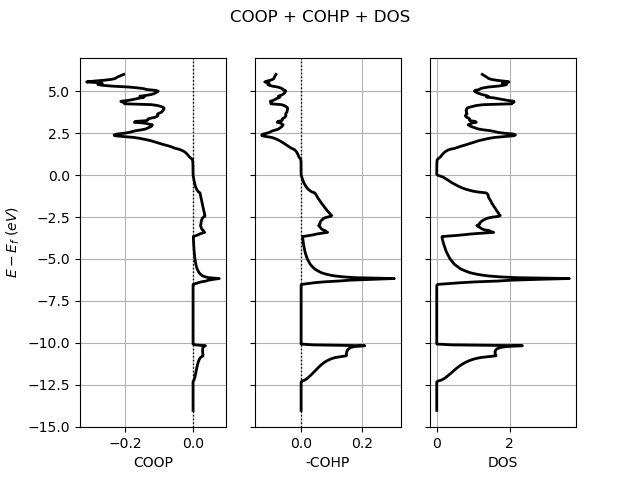
# Plot COOP + COHP + DOS.
lobana.plot(title="COOP + COHP + DOS")
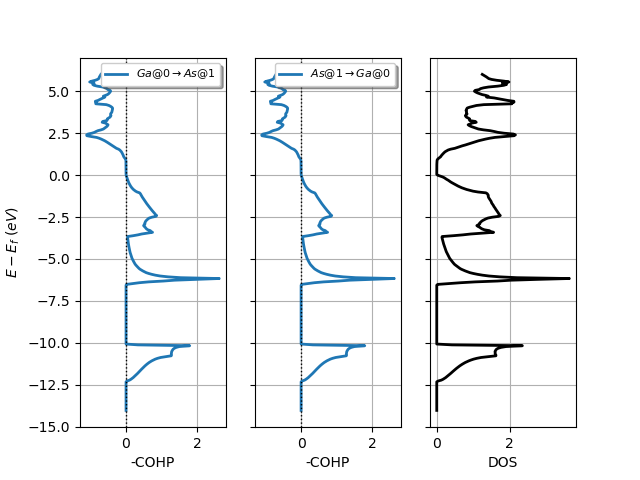
# Plot COHP for all sites in from_site_index and Lobster DOS.
lobana.plot_coxp_with_dos(from_site_index=[0, 1])
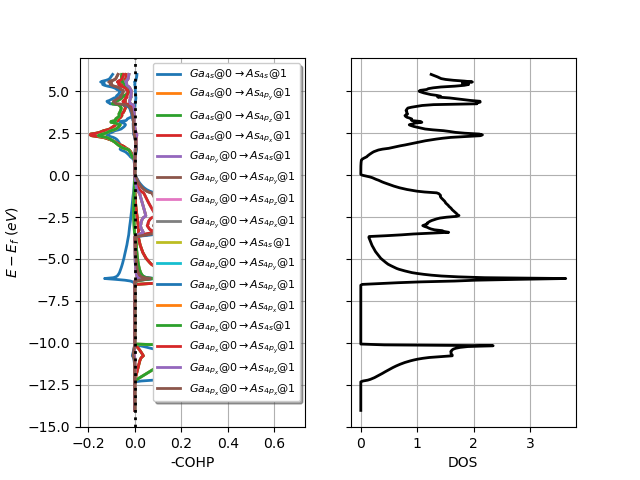
# Plot orbital projections.
lobana.plot_coxp_with_dos(from_site_index=[0], with_orbitals=True)
#lobana.plot_with_ebands(ebands="out_GSR.nc")
Total running time of the script: (0 minutes 0.443 seconds)