Note
Go to the end to download the full example code.
GW Convergence
This example shows how to use the SigresRobot to visualize the convergence of the QP results stored in the SIGRES.nc files produced by the GW code (sigma run).
from abipy.abilab import SigresRobot
import abipy.data as abidata
# List of SIGRES files computed with different values of nband.
filenames = [
"si_g0w0ppm_nband10_SIGRES.nc",
"si_g0w0ppm_nband20_SIGRES.nc",
"si_g0w0ppm_nband30_SIGRES.nc",
]
filepaths = [abidata.ref_file(fname) for fname in filenames]
# Build robot from list of file paths
robot = SigresRobot.from_files(filepaths)
#robot.remap_labels(lambda sigres: sigres.params["sigma_nband"])
# Plot the convergence of the QP gaps.
robot.plot_qpgaps_convergence(sortby="sigma_nband", title="QP gaps vs sigma_nband")
# Plot the convergence of the QP energies.
robot.plot_qpdata_conv_skb(spin=0, kpoint=[0, 0, 0], band=3, sortby="sigma_nband")
# sphinx_gallery_thumbnail_number = 3
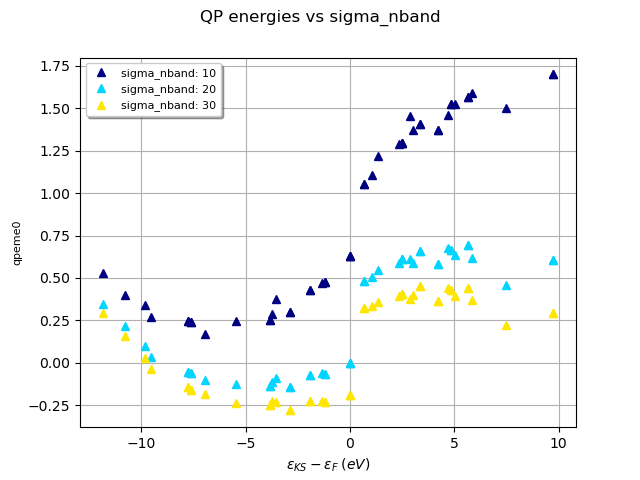
robot.plot_qpfield_vs_e0("qpeme0", sortby="sigma_nband", title="QP energies vs sigma_nband")
Total running time of the script: (0 minutes 0.889 seconds)
![QP gaps vs sigma_nband, k-point: [-0.250, -0.250, +0.000], k-point: [-0.250, +0.250, +0.000], k-point: X [+0.500, +0.500, +0.000], k-point: W [-0.250, +0.500, +0.250], k-point: L [+0.500, +0.000, +0.000], k-point: $\Gamma$ [+0.000, +0.000, +0.000]](../_images/sphx_glr_plot_qpconvergence_001.png)
![QP results spin: 0, k:$\Gamma$ [+0.000, +0.000, +0.000], band: 3](../_images/sphx_glr_plot_qpconvergence_002.png)